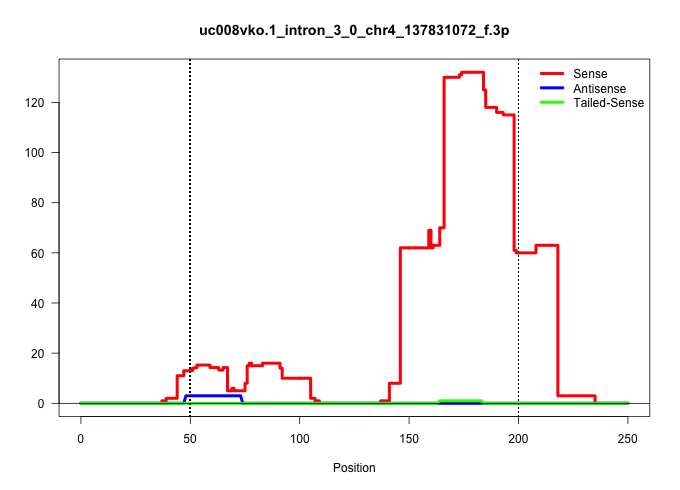
| Gene: MmKIF17 | ID: uc008vko.1_intron_3_0_chr4_137831072_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| GACTGGGGCCACAGGGGAAAGGCTGAAGGAGGCCACCAAAATTAACCTGTCACTGTCGGCGCTGGGCAACGTCATCTCGGCACTGGTGGATGGGCGCTGCAAGCACATTCCTTACCGGGACTCGAAGCTGACCCGACTGCTTCAGGACTCACTGGGTGGCAACACCAAGACTCTGATGGTGGCTTGCCTGTCTCCCGCAGACAACAATTACGATGAGACCCTCAGTACGCTGCGCTATGCTAACAGGGCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................AAGACTCTGATGGTGGCTTGCCTGTCTCCCGCAGACAACAATTACGATGAGA................................ | 52 | 1 | 60.00 | 60.00 | 60.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................ACTCACTGGGTGGCAACACCAAGACTCTGATGGTGGCTTGCCTGTCTCCCGC.................................................... | 52 | 1 | 54.00 | 54.00 | 54.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ACCTGTCACTGTCGGCGCTGGGC....................................................................................................................................................................................... | 23 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TCGGCACTGGTGGATGGGCGCTGCAAGCA................................................................................................................................................. | 29 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAGGACTCACTGGGTGGC.......................................................................................... | 19 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CAACACCAAGACTCTGATGGTGGCTT................................................................. | 26 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCAAGACTCTGATGGTGGCT.................................................................. | 20 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CTCGGCACTGGTGGATG.............................................................................................................................................................. | 17 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TACGATGAGACCCTCAGTACGCTGCGC............... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ....................................................................................................................................................................CCAAGACTCTGATGGTGGCg.................................................................. | 20 | g | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGTCACTGTCGGCGCTGGGCAACGTCATC.............................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGTCGGCGCTGGGCAACGTCATCTC............................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................GATGGTGGCTTGCCTGTCT......................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGATGGTGGCTTGCCTGTCTCCCGCA................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................CGGCACTGGTGGATGGGCGCTGCAAGCACA............................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................AATTAACCTGTCACTGTCGG............................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................AAAATTAACCTGTCACTGTCGGCGCT........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CGTCATCTCGGCACTGGTGGAT............................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................TGCTTCAGGACTCACTGGGTGGCAA........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................GCAACGTCATCTCGGCACTGGTGGAT............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACACCAAGACTCTGATGGTGGCTTGCCTG............................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGTCACTGTCGGCGCTGGGCAACGTCAT............................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGGTGGATGGGCGCTGCAAGCACATT............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................ACTGTCGGCGCTGGGCAAC.................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CACCAAGACTCTGATGGTGGCTTGCCTG............................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................CTGTCGGCGCTGGGC....................................................................................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| GACTGGGGCCACAGGGGAAAGGCTGAAGGAGGCCACCAAAATTAACCTGTCACTGTCGGCGCTGGGCAACGTCATCTCGGCACTGGTGGATGGGCGCTGCAAGCACATTCCTTACCGGGACTCGAAGCTGACCCGACTGCTTCAGGACTCACTGGGTGGCAACACCAAGACTCTGATGGTGGCTTGCCTGTCTCCCGCAGACAACAATTACGATGAGACCCTCAGTACGCTGCGCTATGCTAACAGGGCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................GTCACTGTCGGCGCTGGGCAACGTCA................................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |