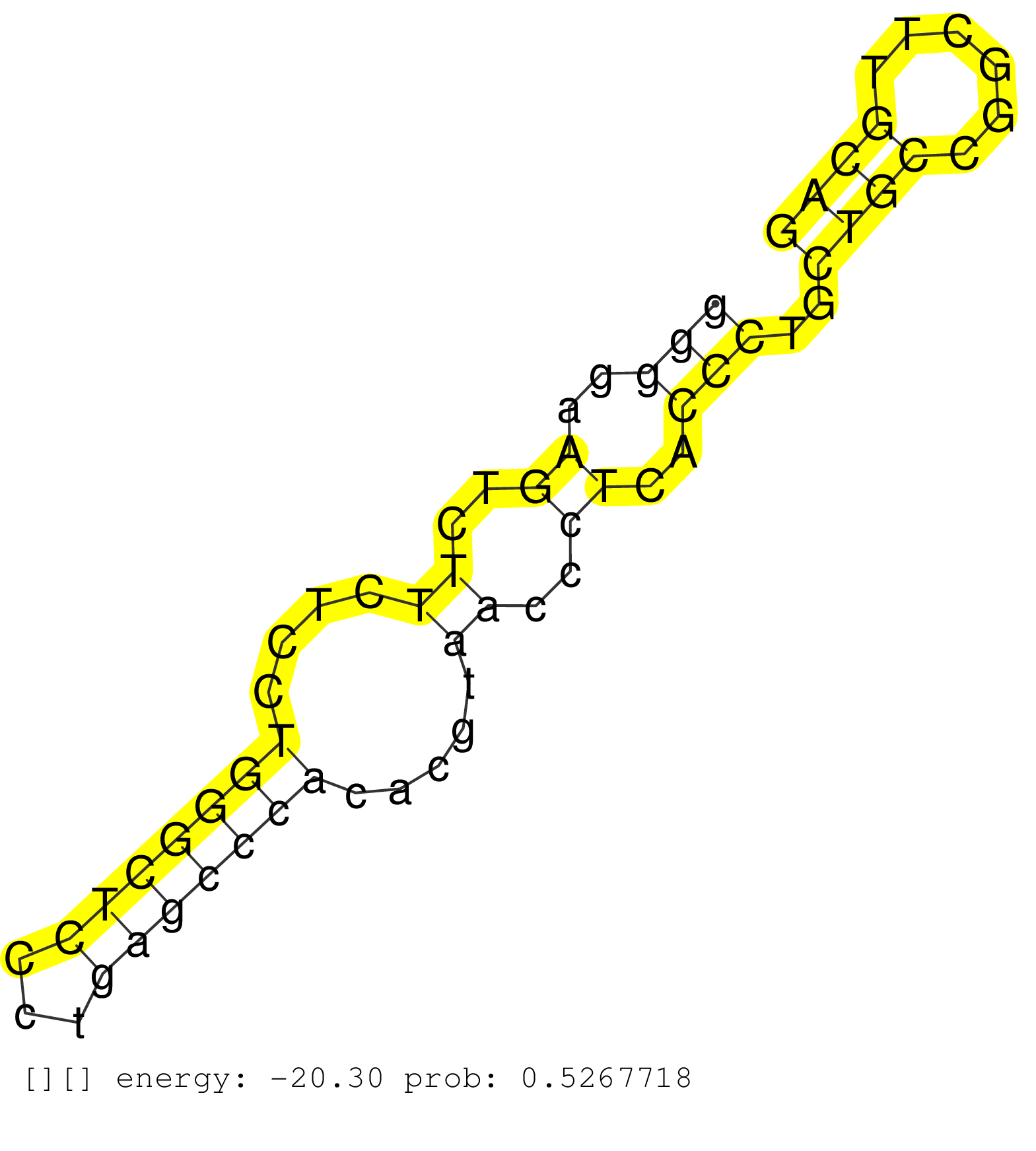

| Gene: Hspg2 | ID: uc008vjc.1_intron_42_0_chr4_137094201_f | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| GTGTGCGGCCCGGAGCTGACGTCACTTTCATCTGTACGGCCAAGAGCAAAGTGAGCAGACAGGGAGTGGTGGGGGAAGTCTTCTCCTGGGCTCCCTGAGCCCACACGTAACCCTCACCCTGCTGCCGGCTTGCAGTCCCCAGCCTACACCCTGGTATGGACCCGTCTGCACAATGGGAAGCTGCC .......................................................................(((..((..((....(((((((...))))))).....))..))..)))..((((......)))).................................................. .......................................................................72.............................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAGT................................................. | 23 | 1 | 20.00 | 20.00 | 11.00 | 5.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTG..................................................... | 19 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAG.................................................. | 22 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAt.................................................. | 22 | t | 3.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CACCCTGCTGCCGGCTTGCAGT................................................. | 22 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CGTCTGCACAATGGGAAGC.... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGa.................................................... | 20 | a | 2.00 | 5.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGC.................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CACCCTGCTGCCGGCTTGCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ACGTCACTTTCATCTGTACGGCCAAGA............................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CTGACGTCACTTTCATCTGTACGGCCAAGAG........................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAa.................................................. | 22 | a | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGTCTTCTCCTGGGCacc........................................................................................... | 18 | acc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGG................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAagt................................................ | 24 | agt | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGGGG.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCcgt................................................. | 23 | cgt | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CCCGGAGCTGACGTCACTTT............................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGGt............................................................................................................... | 24 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAGc................................................. | 23 | c | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GGTATGGACCCGTCTGCACAATGGG........ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CCGGAGCTGACGTCACTTTC............................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTtcag.................................................. | 22 | tcag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCAGa................................................. | 23 | a | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CGGAGCTGACGTCACTTTCATCTGTACGGC................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGGGGttt........................................................................................................... | 28 | ttt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGGGGAtt........................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGCAGACAGGGAGTGGTGGGaac............................................................................................................. | 26 | aac | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............AGCTGACGTCACTTTCATCTGTACGGC................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCACCCTGCTGCCGGCTTGCga.................................................. | 22 | ga | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACCCGTCTGCACAATGGGAAGCTGC. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| GTGTGCGGCCCGGAGCTGACGTCACTTTCATCTGTACGGCCAAGAGCAAAGTGAGCAGACAGGGAGTGGTGGGGGAAGTCTTCTCCTGGGCTCCCTGAGCCCACACGTAACCCTCACCCTGCTGCCGGCTTGCAGTCCCCAGCCTACACCCTGGTATGGACCCGTCTGCACAATGGGAAGCTGCC .......................................................................(((..((..((....(((((((...))))))).....))..))..)))..((((......)))).................................................. .......................................................................72.............................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TTGCAGTCCCCAGCCTACACCCTGGTA............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................GGCTTGCAGTCCCCAGCCTACACCCT................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |