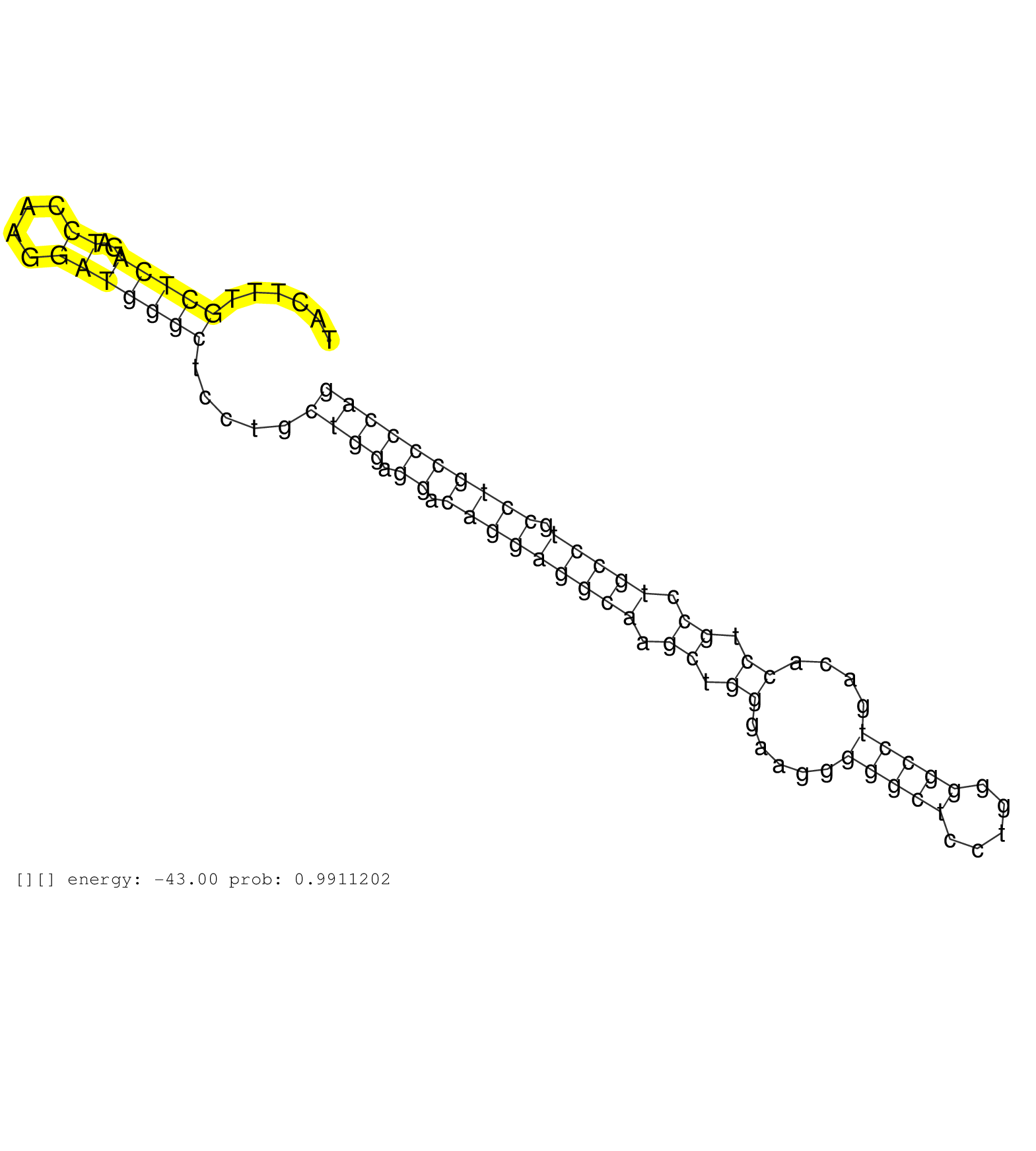

| Gene: Hspg2 | ID: uc008vjc.1_intron_37_0_chr4_137089475_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CCTGAGGATTGCCAGCCCTGTGCCTGCCCGCTGACCAACCCAGAGAACATGTGAGTACTTTGCTCAGATCCAAGGATGGGCTCCTGCTGGAGGACAGGAGGCAAGCTGGGAAGGGGGCTCCTGGGGCCTGACACCTGCCTGCCTGCCTGCCCCAGGTTCTCCCGCACCTGTGAGAGCCTTGGAGCTGGAGGGTACCGCTGCACCG .............................................................(((((..((....))))))).....((((.((.(((((((((.((.((.....(((((.....)))))....)).)).))))).)))))))))).................................................. .......................................................56.................................................................................................155................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTACTTTGCTCAGATCCAAGGAT................................................................................................................................ | 27 | 1 | 22.00 | 22.00 | - | 6.00 | - | 9.00 | - | - | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGAGTACTTTGCTCAGATCCAAGGA................................................................................................................................. | 26 | 1 | 12.00 | 12.00 | - | 5.00 | - | - | - | - | 1.00 | 1.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 |
| ..........................................................................................AGGACAGGAGGCAAGCTGGGA.............................................................................................. | 21 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCTGGAGGACAGGAGGCAAGCTa................................................................................................. | 24 | a | 6.00 | 2.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGAGGACAGGAGGCAAGCTGG................................................................................................ | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AGGATTGCCAGCCCTGTGCCTGC.................................................................................................................................................................................. | 23 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACTTTGCTCAGATCCAAGG.................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCTGGAGGACAGGAGGCAAGCT.................................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCTGGAGGACAGGAGGCAAGCTagaa.............................................................................................. | 27 | agaa | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGAGGACAGGAGGCAAGCTGGa............................................................................................... | 23 | a | 2.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACTTTGCTCAGATCCAAGGATGG.............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................ACCTGTGAGAGCCTTGGAG..................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTACTTTGCTCAGATCCAAGGATG............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCTGGAGGACAGGAGGCAAGa................................................................................................... | 22 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGAGAGCCTTGGAGCTGGAGGGTACCG........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTACTTTGCTCAGATCCAAGGATGGG............................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGAGAGCCTTGGAGCTGGAGGGTACCGCTGCA... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCACCTGTGAGAGCCTTGGAGCTGG................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGAGGACAGGAGGCAAGCTG................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGTGAGAGCCTTGGAGCTGGAGGGTt........... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................TGCCCGCTGACCAACCCAGAGAACATGTt........................................................................................................................................................ | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .CTGAGGATTGCCAGCCCTGTGCCTGC.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCACCTGTGAGAGCCTTGG....................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| CCTGAGGATTGCCAGCCCTGTGCCTGCCCGCTGACCAACCCAGAGAACATGTGAGTACTTTGCTCAGATCCAAGGATGGGCTCCTGCTGGAGGACAGGAGGCAAGCTGGGAAGGGGGCTCCTGGGGCCTGACACCTGCCTGCCTGCCTGCCCCAGGTTCTCCCGCACCTGTGAGAGCCTTGGAGCTGGAGGGTACCGCTGCACCG .............................................................(((((..((....))))))).....((((.((.(((((((((.((.((.....(((((.....)))))....)).)).))))).)))))))))).................................................. .......................................................56.................................................................................................155................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......CCAGCCCTGTGCCTGCagca.................................................................................................................................................................................. | 20 | agca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |