
| Gene: Zbtb40 | ID: uc008vit.1_intron_0_0_chr4_136539160_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(16) TESTES |
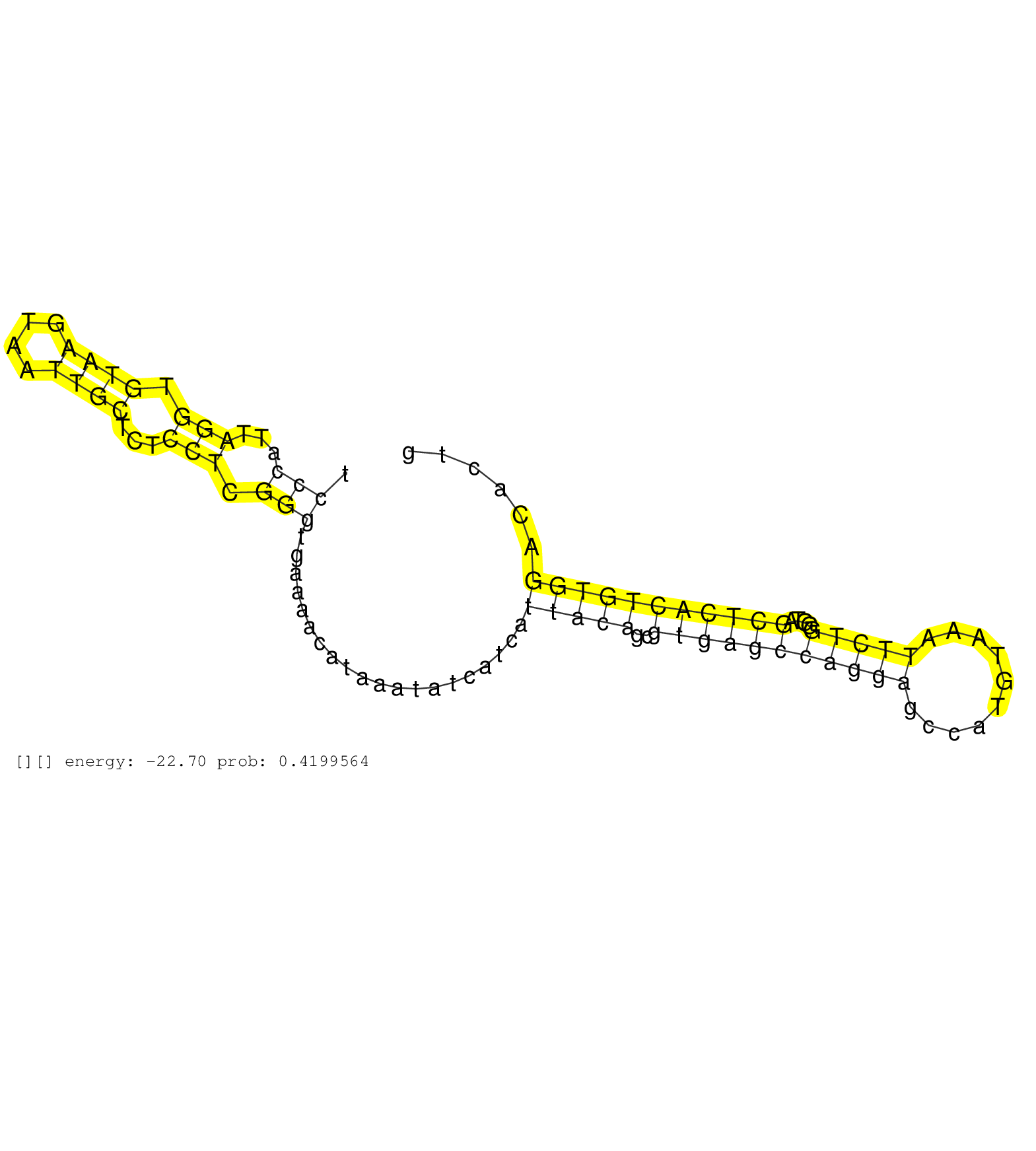
| AGAGTTATTTAAATTGTCTGCCTGCCTCTCTTTTGCATCGCTGGTGTATGAAACTTCCCTCTTCCCATTAGGTGTAAGTAATTGCTCTCCTCGGGTGAAAACATAAATATCATCATTACAGCGTGAGCCAGGAGCCATGTAAATTCTGGGTAGCTCACTGTGGACACTGCCCAGAGCAGCCTTCCTGTCTTTTCCTCCAGGTGATCCGAGCTCCGGAGCCAGCGGCCCCGGCCGAGCAGGTGATCACGTT ...............................................................(((...(((.((((....))))...))).)))....................(((((..(((((((((((..........)))))....)))))))))))....................................................................................... ..............................................................63........................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................TGTAAATTCTGGGTAGCTCACTGTGGAC..................................................................................... | 28 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCACTGTGGACACTGCCCAGAGCAGCCTT................................................................... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CACTGTGGACACTGCCCAGAGCAGCCTTCC................................................................. | 30 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................TGTAAATTCTGGGTAGCTCACTGTGG....................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCACTGTGGACACTGCCCAGAGCAGCCT.................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGAGCCAGGAGCCATGTAAATTCTGGGT................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................TCGCTGGTGTATGAAACTTCCCTCTTCCC........................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAAATATCATCATTACAGCGTGAGCCA........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TCTTTTCCTCCAGGTGATCCGAGCTC..................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCACTGTGGACACTGCCCAGAGCAGCC..................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CACTGTGGACACTGCCCAGAGCAGCCT.................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTCCTGTCTTTTCCTCCAGGTGATCCGAGC....................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGCCTCTCTTTTGCATCGCTGGTGTAT......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TATCATCATTACAGCGTGAGCCAGGAG.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAAATTCTGGGTAGCTCACTGTGG....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................TGCCTGCCTCTCTTTTGCATCGCTGG.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGGGTAGCTCACTGTGGACACTGCCCAG............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........TAAATTGTCTGCCTGCCTCTCTTTTGCATC................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTCCTGTCTTTTCCTCCAGGTGAT............................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................TCACTGTGGACACTGCCCAGAGCAGCCTTC.................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CACTGTGGACACTGCCCAGAGCAGCCTT................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTAGGTGTAAGTAATTGCTCTCCTCGG............................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGTAAATTCTGGGTAGCTCACTGTGt....................................................................................... | 26 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TCCCATTAGGTGTAAGTAATTGCTCTC................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ATCATTACAGCGTGAGCCAGGAGCCA................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTCCCATTAGGTGTAAGTAATTGCTCTC................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGTCTTTTCCTCCAGGTGATCCGAGC....................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................TAAATATCATCATTACAGCGTGAGCCAG....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................TGAAACTTCCCTCTTCCCATTAGGTGT............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| AGAGTTATTTAAATTGTCTGCCTGCCTCTCTTTTGCATCGCTGGTGTATGAAACTTCCCTCTTCCCATTAGGTGTAAGTAATTGCTCTCCTCGGGTGAAAACATAAATATCATCATTACAGCGTGAGCCAGGAGCCATGTAAATTCTGGGTAGCTCACTGTGGACACTGCCCAGAGCAGCCTTCCTGTCTTTTCCTCCAGGTGATCCGAGCTCCGGAGCCAGCGGCCCCGGCCGAGCAGGTGATCACGTT ...............................................................(((...(((.((((....))))...))).)))....................(((((..(((((((((((..........)))))....)))))))))))....................................................................................... ..............................................................63........................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TTCCCTCTTCCCATTAaaaa.................................................................................................................................................................................... | 20 | aaaa | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................AGCAGCCTTCCTGTggaa.............................................................. | 18 | ggaa | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ......................................................................................................................................................................................TGTCTTTTCCTCCAGGTtca................................................ | 20 | tca | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |