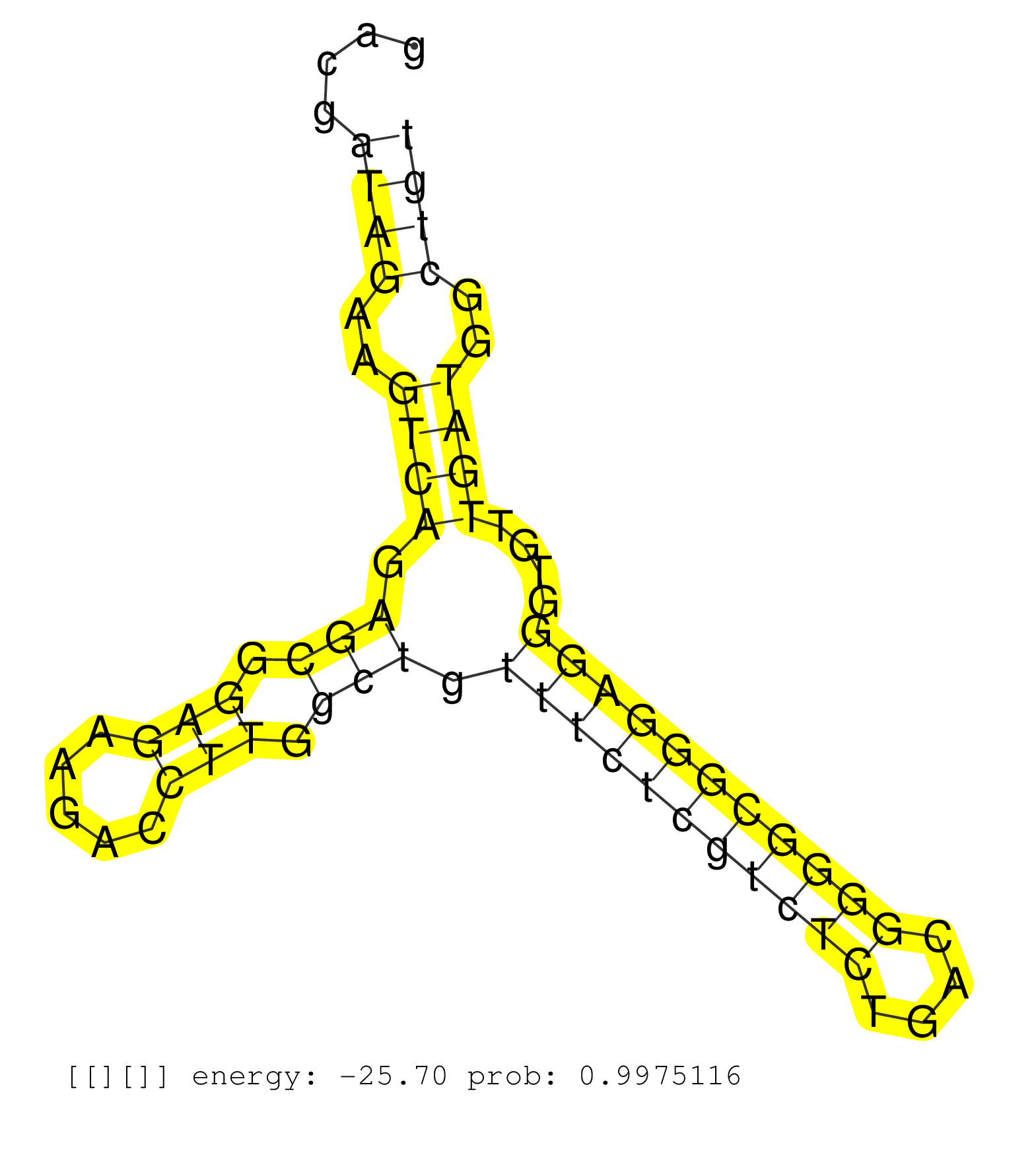

| Gene: Rpl11 | ID: uc008vhr.1_intron_4_0_chr4_135608633_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| AGTGTCTGTTCCCTACTCGTGGATCCGCTTGGGATTCAGGCTCTCTCTCTGTGTTCGTTCTCTTCCAAACGCGGAGACCTTGGTGGTCCCGAGGGCTTAGGCGGGATCCGGGACGATAGAAGTCAGAGCGGAGAAGACCTTGGCTGTTTCTCGTCTCTGACGGGGCGGGAGGGTGTTGATGGCTGTGTCCCTCTTCGCAGCAAGATCAAGGGGAAAAGGAGAACCCCATGCGGGAACTGCGCATCCGCAA ...................................................................................................................((((..((((.(((.(((.....))).))).(((((((((((....)))))))))))....))))..))))................................................................ ...............................................................................................................112.......................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................TAGAAGTCAGAGCGGAGAAGACCTTG............................................................................................................ | 26 | 1 | 8.00 | 8.00 | - | 2.00 | - | 1.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................TAGAAGTCAGAGCGGAGAAGACCTT............................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................TCAGGCTCTCTCTCTGTGTTCGTTCTC............................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTAGGCGGGATCCGGGACGATAGAA................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGGCTGTTTCTCGTCTCTGACGGGGC.................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGCGGGATCCGGGACGATAGAAGT............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TCAGGCTCTCTCTCTGTGTTCGTTCa............................................................................................................................................................................................. | 26 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................GAAGACCTTGGCTGTTTCTCGTCTCTGA.......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATCCGGGACGATAGAAGTCAGAGCGGA...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TTCGCAGCAAGATCAAGGGGAAAAGGAG............................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGACGGGGCGGGAGGGTGTTGATGt.................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TTCAGGCTCTCTCTCTGTGTTCGTTCT............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCGCAGCAAGATCAAGGGGAAAAGGAG............................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GATAGAAGTCAGAGCGGAGAAGACCTTGGCTGT....................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGAAGTCAGAGCGGAGAAGACCTTGGC.......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................TTCGCAGCAAGATCAAGGGGAAAAGGAGAACC......................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGAAGTCAGAGCGGAGAAGACCTTGG........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CGATAGAAGTCAGAGCGGAGAAGACCT.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGAAGTCAGAGCGGAGAAGACatt............................................................................................................. | 25 | att | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGGGATTCAGGCTCTCTCTCTGTGTT................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GATGGCTGTGTCCCTCTTCGCAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TCTCTGTGTTCGTTCTCTTCCAAACGC.................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGAAGTCAGAGCGGAGAAGACC............................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ATGCGGGAACTGCGCATa..... | 18 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................CAAGATCAAGGGGAAAAGGAGAAC.......................... | 24 | 6 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCAAGGGGAAAAGGAGAACCCCATGC................... | 26 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| .......................................................................................................................................................................................................................AAGGAGAACCCCATGCGGGAACTGCGCAT...... | 29 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| .....................................................................................................................................................................................................................AAAAGGAGAACCCCATGCGGGAACTGCG......... | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| ......................................................................................................................................................................................................................................CGGGAACTGCGCATCCGCAA | 20 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GGGGAAAAGGAGAACCCCATGCGGGA............... | 26 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AAGGGGAAAAGGAGAACCCCAT..................... | 22 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| .......................................................................................................................................................................................................GCAAGATCAAGGGGAAAAGGAGAAC.......................... | 25 | 6 | 0.17 | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGTGTCTGTTCCCTACTCGTGGATCCGCTTGGGATTCAGGCTCTCTCTCTGTGTTCGTTCTCTTCCAAACGCGGAGACCTTGGTGGTCCCGAGGGCTTAGGCGGGATCCGGGACGATAGAAGTCAGAGCGGAGAAGACCTTGGCTGTTTCTCGTCTCTGACGGGGCGGGAGGGTGTTGATGGCTGTGTCCCTCTTCGCAGCAAGATCAAGGGGAAAAGGAGAACCCCATGCGGGAACTGCGCATCCGCAA ...................................................................................................................((((..((((.(((.(((.....))).))).(((((((((((....)))))))))))....))))..))))................................................................ ...............................................................................................................112.......................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................TCTCTCTCTGTGTTCGTTCTCTTCCA....................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................GGATCCGCTTGGGATTCAGGCTCTCTC........................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................GCTCTCTCTCTGTGTTCGTTCTCTTCCA....................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGACCTTGGCTGTTTCTCGTCTCT............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................AGGCTCTCTCTCTGTGTTCGTTCTCTTCCAt....................................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |