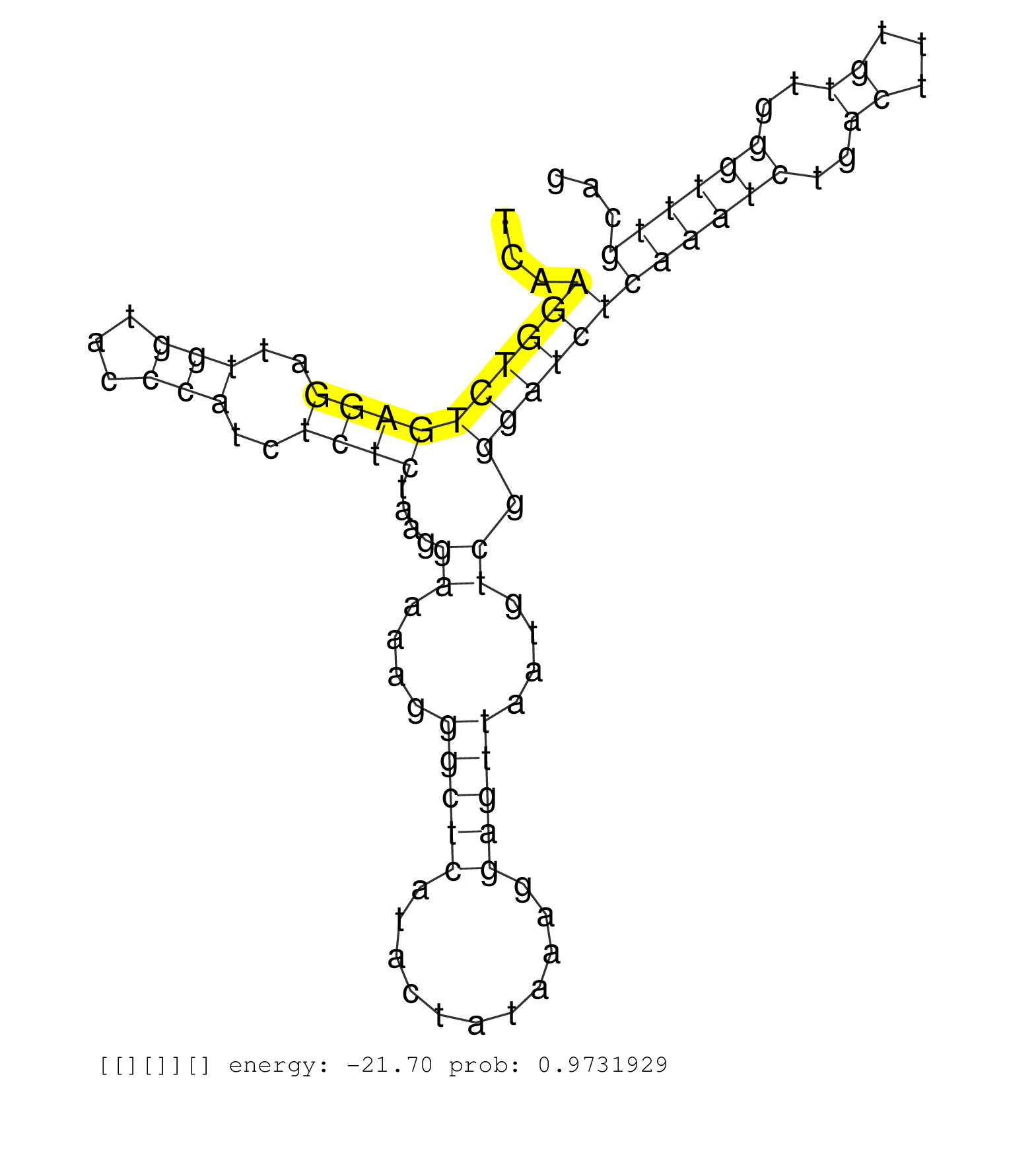
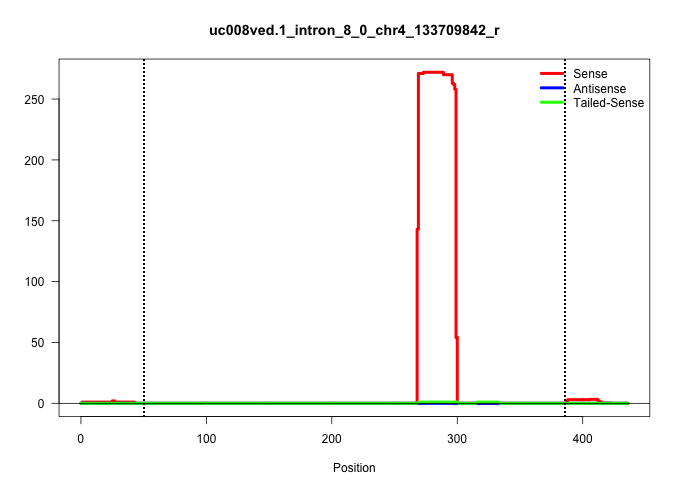
| Gene: Ccdc21 | ID: uc008ved.1_intron_8_0_chr4_133709842_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(24) TESTES |
| CTGAACAGTAACGAGCATCTCCTGAAGGAGAAGGAGCTCCTCATTGACAAGTAAGATGGCGGGGCCCGTGCCAGGGCAGTGACCTCTTTCCTCGGGGTAGAGCAGCTCGGTCTTGCAGGGCTTGGGGGTGGGCGAGTGATTCATTGCAGCTCATCTTTCCTTGGTTTTTCCCATTAAACTTTTTCTCATTCGCTCCTACCTTTTCAGCAAAATATAATGTCCCACATACTTTTGGGGCATAGTGGTAGAGCATACATAGCTGCCTTCCAAACTGTGATGTCTAGAATCAAGGTCTGAGGATTGGTACCCATCTCTCTAAGGAAAAGGGCTCATACTATAAAGGAGTTAATGTCGGGATCTCAAATCTGACTTTGTTGGGTTTGCAGGCAAAGGAAGCATATCTCTCAGCTGGAGCAGAAAGTGCGAGAGAGTGAAC .................................................................................................................................................................................................................................................................................................((((((((((..(((...)))..))))....((....(((((...........)))))....)).))))))((((((..((...))..))))))..................................................... ..............................................................................................................................................................................................................................................................................................287................................................................................................386................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTGAGG......................................................................................................................................... | 30 | 1 | 113.00 | 113.00 | 44.00 | 39.00 | 12.00 | 2.00 | 3.00 | 4.00 | - | 3.00 | - | 1.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTGAGG......................................................................................................................................... | 31 | 1 | 108.00 | 108.00 | 38.00 | 41.00 | 12.00 | 5.00 | 2.00 | 1.00 | - | - | 1.00 | - | 2.00 | - | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTGAGGA........................................................................................................................................ | 32 | 1 | 49.00 | 49.00 | 15.00 | 22.00 | 2.00 | - | 2.00 | - | 4.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTGAGGA........................................................................................................................................ | 31 | 1 | 12.00 | 12.00 | 7.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTG............................................................................................................................................ | 28 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTGAG.......................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTG............................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTGAGGATa...................................................................................................................................... | 34 | a | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCA................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................TAAGGAAAAGGGCTgcac...................................................................................................... | 18 | gcac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................CAAAGGAAGCATATCTCTCAGCTGG........................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTGA........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTGAGGATTG..................................................................................................................................... | 35 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................GGAAGCATATCTCTCAGCTGGAGCAGAAAGTG............. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................................................................................AACTGTGATGTCTAGAATCAAGGTCTGAGGAa....................................................................................................................................... | 32 | a | 1.00 | 12.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................AAACTGTGATGTCTAGAATCAAGGTCTGAG.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................AAAGGAAGCATATCTCTCAGCTGGA....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................AGGAGAAGGAGCTCCTCA......................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................GTGATGTCTAGAATCAAGGTCTGAGGA........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGAACAGTAACGAGCATCTCCTGAAG......................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................CAAAGGAAGCATATCTCTCAGCTGGAGC..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TAAGATGGCGGGGCagaa............................................................................................................................................................................................................................................................................................................................................................................... | 18 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................AGCTGGAGCAGAAAGTG............. | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - |
| CTGAACAGTAACGAGCATCTCCTGAAGGAGAAGGAGCTCCTCATTGACAAGTAAGATGGCGGGGCCCGTGCCAGGGCAGTGACCTCTTTCCTCGGGGTAGAGCAGCTCGGTCTTGCAGGGCTTGGGGGTGGGCGAGTGATTCATTGCAGCTCATCTTTCCTTGGTTTTTCCCATTAAACTTTTTCTCATTCGCTCCTACCTTTTCAGCAAAATATAATGTCCCACATACTTTTGGGGCATAGTGGTAGAGCATACATAGCTGCCTTCCAAACTGTGATGTCTAGAATCAAGGTCTGAGGATTGGTACCCATCTCTCTAAGGAAAAGGGCTCATACTATAAAGGAGTTAATGTCGGGATCTCAAATCTGACTTTGTTGGGTTTGCAGGCAAAGGAAGCATATCTCTCAGCTGGAGCAGAAAGTGCGAGAGAGTGAAC .................................................................................................................................................................................................................................................................................................((((((((((..(((...)))..))))....((....(((((...........)))))....)).))))))((((((..((...))..))))))..................................................... ..............................................................................................................................................................................................................................................................................................287................................................................................................386................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................................................................................................................GTTGGGTTTGCAGGCAgggt............................................... | 20 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |