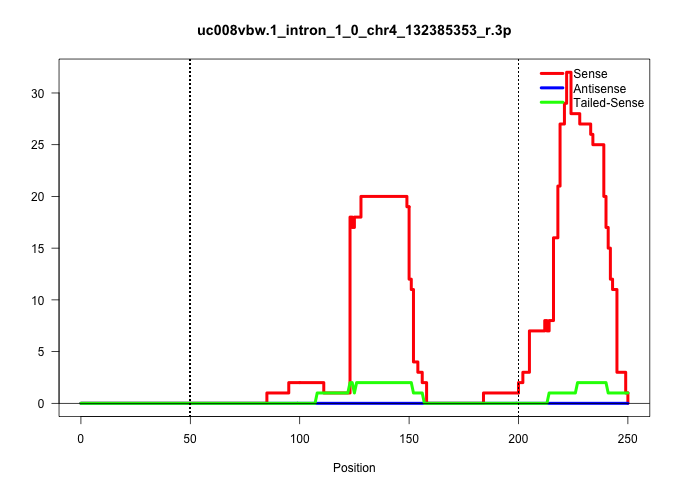
| Gene: Ppp1r8 | ID: uc008vbw.1_intron_1_0_chr4_132385353_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
| CAGTGCATCACTGTGTGAGACAGTGATGTCTGCTGCTTGAGCTGCCTTGCTCTGCTCCTCGTGTACTCGGCGCTGTTGGAGCCGTTGGGAAGGCTTAGCACATACGATCCCTAGCATTAGCTATGTTTCCTGTTTGAACGGGCTGACGGCTCTTTGGCTTGATGACCCCATATTCTCCCAACCTTGCTCCGTCTCTGCAGAGGACGTGGATCCGTCTGTTGGTCGGTTCCGGAACATGGTGCAGACAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGC.................................................................................................... | 27 | 1 | 7.00 | 7.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGGTCGGTTCCGGAACATGGTGCAGA..... | 26 | 1 | 6.00 | 6.00 | 1.00 | - | - | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGCTC.................................................................................................. | 29 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TTGGTCGGTTCCGGAACATGG........... | 21 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GTGGATCCGTCTGTTGGTC.......................... | 19 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGTTGGTCGGTTCCGGAACATGGTGC........ | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGTTGGTCGGTTCCGGAACATGGTG......... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................GTCGGTTCCGGAACATGGTGCAGA..... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TCGGTTCCGGAACATGGTGCAGACAGC. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGTTGGTCGGTTCCGGAACATGGT.......... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................CCTGTTTGAACGGGCTGACGGCTCTTTGGC............................................................................................ | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTCCTGTTTGAACGGGCTGACGGCTCTTTGt............................................................................................. | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGCTCTTTG.............................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGCT................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TAGCACATACGATCCCTAGCATTAGCTAT.............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGG..................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGCTt.................................................................................................. | 29 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGCTCCGTCTCTGCAGAGGACGTGGATCC..................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGTTGGTCGGTTCCGGAACATGGTGCA....... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CGTCTGTTGGTCGGTTCCGGA................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCTGTTGGTCGGTTCCGGAACATGGTt......... | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TCCGGAACATGGTGCAGACAGCAtt | 25 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................TGGGAAGGCTTAGCACATACGATCCC........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGTTTCCTGTTTGAACGGGCTGACGGCTCTT................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................TCTGTTGGTCGGTTCCGGAACATGGT.......... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AGGACGTGGATCCGTCTGTTGGTCGGTTCCGGAA................ | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CCCTAGCATTAGCTctt............................................................................................................................. | 17 | ctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................................TCGGTTCCGGAACATGGTGCAGACAGCA | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GACGTGGATCCGTCTGTTGGTCGGTT...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TTTCCTGTTTGAACGGGCTGACGGCTC.................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGTGCATCACTGTGTGAGACAGTGATGTCTGCTGCTTGAGCTGCCTTGCTCTGCTCCTCGTGTACTCGGCGCTGTTGGAGCCGTTGGGAAGGCTTAGCACATACGATCCCTAGCATTAGCTATGTTTCCTGTTTGAACGGGCTGACGGCTCTTTGGCTTGATGACCCCATATTCTCCCAACCTTGCTCCGTCTCTGCAGAGGACGTGGATCCGTCTGTTGGTCGGTTCCGGAACATGGTGCAGACAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|