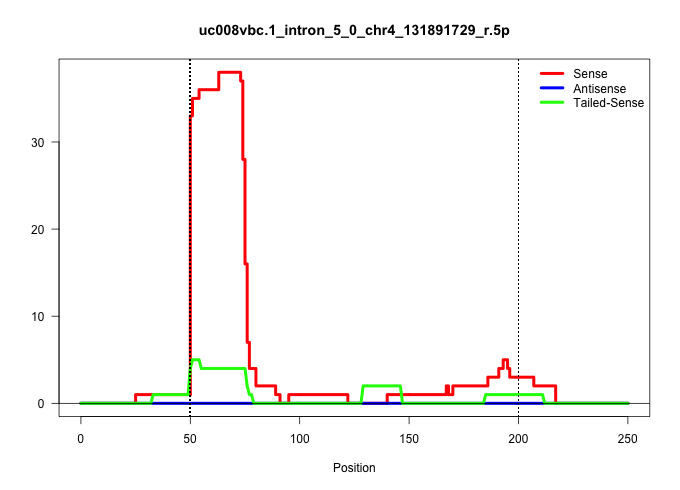
| Gene: Rcc1 | ID: uc008vbc.1_intron_5_0_chr4_131891729_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(18) TESTES |
| CAGCAGCCCTCACCGAAGACGGCCGTGTCTTCCTTTGGGGCTCTTTCCGGGTAAGACTGGGGCTGGCAGGCAGAAATTAACTCAGGATCCCCAAATGATCCACATGCATGGGTAGGGCCCGGATTCTAATCCCTGGCATCTTCGTATGGGCCTGTCCACTCCACGGCCTATGTGCTTGAGAGTGATGCATGAATTGTGTGTCTTTGGGAAGGGATGGGTCAGGGTTGCCTCTGGGCTGAGCTGGCCTTGG ..........................................................((((.((.((((.((((.........(((((.......))))).......((((.....))))..))))....)))).)).))))........................................................................................................... ..................................................51..................................................................................................151................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAA............................................................................................................................................................................... | 25 | 1 | 12.00 | 12.00 | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGA................................................................................................................................................................................ | 24 | 1 | 9.00 | 9.00 | 5.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAAA.............................................................................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 2.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAAAT............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCCTGGCATCTTCacca....................................................................................................... | 18 | acca | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAAt.............................................................................................................................................................................. | 26 | t | 2.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................AATTGTGTGTCTTTGGGAAGGGATGG................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAAAa............................................................................................................................................................................. | 27 | a | 1.00 | 8.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CTATGTGCTTGAGAGTGATGCATGAATT....................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAG................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TAAGACTGGGGCTGGCAGGCAGAAA.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TTCGTATGGGCCTGTCCACTCCACGGCC.................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................GCATGAATTGTGTGTCTTTGG........................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................TTTGGGGCTCTTTCCGGGacaa................................................................................................................................................................................................... | 22 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGACTGGGGCTGGCAGGCAGAAtaat........................................................................................................................................................................... | 28 | taat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGATCCACATGCATGGGTAGGGCCCGG................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGCAGGCAGAAATTAACTCAGGATC................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GACTGGGGCTGGCAGGCAGAAATTAA.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGCATGAATTGTGTGTCTTTGGGAAGt...................................... | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGACTGGGGCTGGCAGGCAGAAAT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGCAGGCAGAAATTAACTCAGGATCCC............................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGACTGGGGCTGGCAGGCAGAAATTAA.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................TGTGCTTGAGAGTGATGCATGAATTG...................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................TGTCTTCCTTTGGGGCTCTTTCCGG........................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................TTGTGTGTCTTTGGGAAGGGATGG................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGCAGCCCTCACCGAAGACGGCCGTGTCTTCCTTTGGGGCTCTTTCCGGGTAAGACTGGGGCTGGCAGGCAGAAATTAACTCAGGATCCCCAAATGATCCACATGCATGGGTAGGGCCCGGATTCTAATCCCTGGCATCTTCGTATGGGCCTGTCCACTCCACGGCCTATGTGCTTGAGAGTGATGCATGAATTGTGTGTCTTTGGGAAGGGATGGGTCAGGGTTGCCTCTGGGCTGAGCTGGCCTTGG ..........................................................((((.((.((((.((((.........(((((.......))))).......((((.....))))..))))....)))).)).))))........................................................................................................... ..................................................51..................................................................................................151................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................GCTGAGCTGGCCTTgagg.. | 18 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |