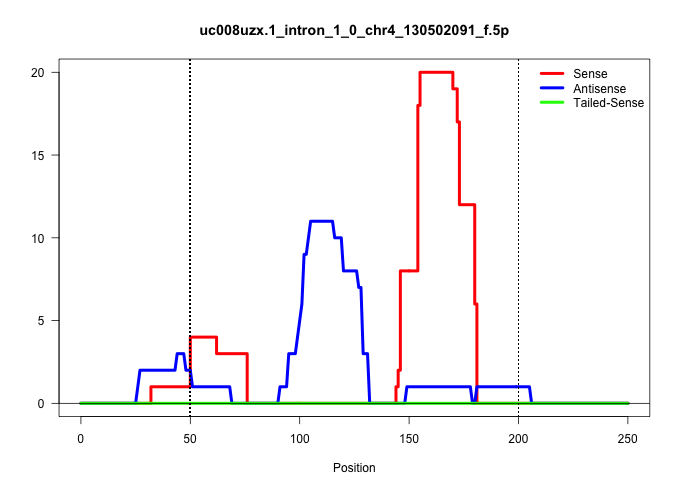
| Gene: Matn1 | ID: uc008uzx.1_intron_1_0_chr4_130502091_f.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(12) TESTES |
| CCTTGAGTGATGCTGAGGGTGGACGCGCCAGATCCCCCGACATCAGCAAGGTATGGAGCACGGGATGGTTGGAGCCTGGGATGAAGCCATAGGGTCCTCCTGGCCTCTCTGTTCAGAGCAGCTGTTTTAGGAACGAGAGAGTCATTTGCAAACATTACTCTGTAGAAGTGGCGGTGGTAGGGTTGAGATCTTCCCTCGAGCAGAGAGAGGCGGGCCAGGTGCTTATGCTAATACTGGCCACCGAGAGAAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................TTACTCTGTAGAAGTGGCGGTGGTAG...................................................................... | 26 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGCAAACATTACTCTGTAGAAGTGGCG............................................................................. | 27 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTACTCTGTAGAAGTGGCGGTGGTAGG..................................................................... | 27 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGGAGCACGGGATGGTTGGAGCC.............................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TACTCTGTAGAAGTGGCGGTGGTAGG..................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TGCAAACATTACTCTGTAGAAGTGGC.............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TTTGCAAACATTACTCTGTAGAAGTGGC.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTGCAAACATTACTCTGTAGAAGTG................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................TCCCCCGACATCAGCAAGGTATGGAGCACG............................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| CCTTGAGTGATGCTGAGGGTGGACGCGCCAGATCCCCCGACATCAGCAAGGTATGGAGCACGGGATGGTTGGAGCCTGGGATGAAGCCATAGGGTCCTCCTGGCCTCTCTGTTCAGAGCAGCTGTTTTAGGAACGAGAGAGTCATTTGCAAACATTACTCTGTAGAAGTGGCGGTGGTAGGGTTGAGATCTTCCCTCGAGCAGAGAGAGGCGGGCCAGGTGCTTATGCTAATACTGGCCACCGAGAGAAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................CCTCCTGGCCTCTCTGTTCAGAGCA.................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GCCTCTCTGTTCAGAGCAGCTGTTTTA......................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - |
| ...........................................................................................GGGTCCTCCTGGCCTCTCTGTTCAG...................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................AGCAAGGTATGGAGCACGGGATGGT..................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGGCCTCTCTGTTCAGAGCAGCTGTTTTA......................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................CTGGCCTCTCTGTTCAGAGCAGCTGTTTTAGGA...................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................GCCTCTCTGTTCAGAGCAGCTGTTT........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................CTCTCTGTTCAGAGCAGCTGTTTTAGGA...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................TCTCTGTTCAGAGCAGCTGTTTTAGGA...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................TTGGAGCCTGGGATGAgac...................................................................................................................................................................... | 19 | gac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................CCAGATCCCCCGACATCAGCA.......................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................GTTGAGATCTTCCCTCGAGCAGAGA............................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................GGCCTCTCTGTTCAGAGCAGCTGTTTTA......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................GCCAGATCCCCCGACATCAGCAAGG....................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AAACATTACTCTGTAGAAGTGGCGGTGGTA....................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |