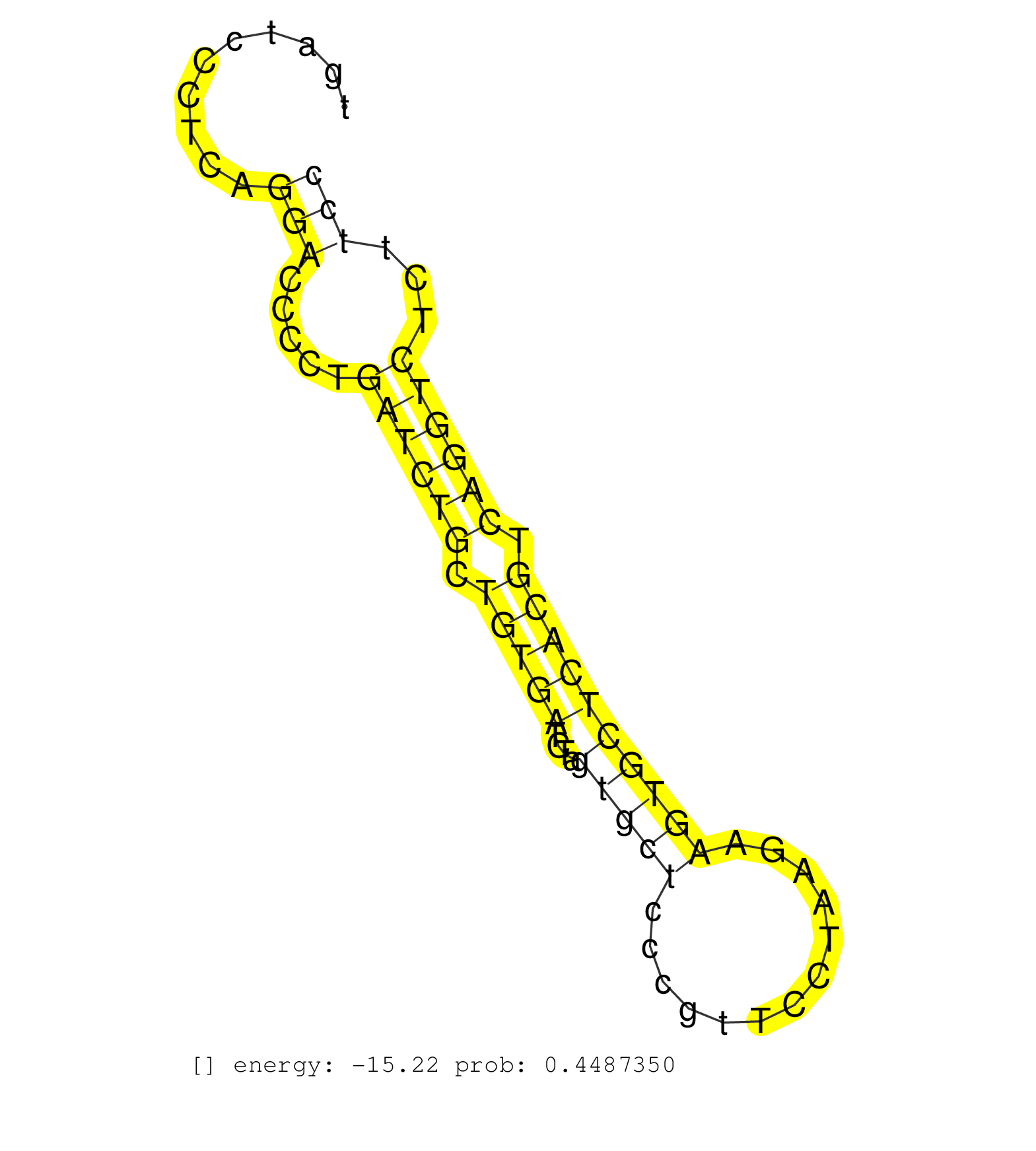

| Gene: Pef1 | ID: uc008uyw.1_intron_3_0_chr4_129803042_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| AGCCTTGATAGTGAAGAGATGGGAGGCAGGGCACACCCAAAGAAATCTCCTGTGCTCCTGATGTCTGACTGGCTCCCCAAGACTTCTCTGTCCTCATGGTTCTGAGCTGATCCCTCAGGACCCCTGATCTGCTGTGATCTAGTGCTCCCGTTCCTAAGAAGTGCTCACGTCAGGTCTCTTCCTGTGCCCTTCCCCTCCAGCGCTCTCCCAGATGGGCTACAACCTGAGCCCTCAGTTCACGCAGCTCCTG .....................................................................................................................(((.....((((((.(((((....(((((.............)))))))))).))))))...))).................................................................... ...........................................................................................................108.......................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................TCCTAAGAAGTGCTCACGTCAGGTCTC........................................................................ | 27 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ACAACCTGAGCCCTCAGTTCACGCAGCT.... | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................ACAACCTGAGCCCTCAGTTCACGC........ | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................TAAGAAGTGCTCACGTCAGGTCTCTT...................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CTCTTCCTGTGCCCTTCCCCTCCAGC................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGATGGGCTACAACCTGAGCC.................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AACCTGAGCCCTCAGTTCACGCAGCTCC.. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCTCAGGACCCCTGATCTGCTGTGATCT.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGTGCTCCCGTTCCTAAGAAGTactc.................................................................................... | 27 | actc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCTGCTGTGATCTAGTGCTCCCGTa.................................................................................................. | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........TAGTGAAGAGATGGGAGGCAGGGCA......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ACAACCTGAGCCCTCAGTTCACGCAGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGCGCTCTCCCAGATGGGCTtt.............................. | 22 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................TGTCCTCATGGTTCTGAGCTGATCCCT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CTCTTCCTGTGCCCTTCCCCTCCAG.................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................ACCTGAGCCCTCAGTTCACGCAGCTCC.. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTGATCTAGTGCTCCCGTTCCTAcg............................................................................................ | 26 | cg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGATCTAGTGCTCCCGTTCCTAAGAAGT........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................TAGTGCTCCCGTTCCTAAGAAGTGC...................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCTGCTGTGATCTAGTGCTCCCGTgcc................................................................................................ | 27 | gcc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ACAACCTGAGCCCTCAGTTCACGCAGCTCCT. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCTGCTGTGATCTAGTGCTCCCGTTC................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGTGCTCCCGTTCCTAAGAAGTGCTC.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TAGTGAAGAGATGGGAGGCAGGGCACACC..................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TAAGAAGTGCTCACGTCAGGTCTCTTCC.................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CTGATCCCTCAGGACCCCTGATCTG....................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TAGTGAAGAGATGGGAGGCAGGGC.......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGTGCTCCTGATGTCTGACTGGCTCC.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGAGCCCTCAGTTCACGCAGCTCCTG | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................ACAACCTGAGCCCTCAGTTCACGCAGCTCC.. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGATGTCTGACTGGCTCCCCAAGAC....................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GAAGTGCTCACGTCgcgg........................................................................... | 18 | gcgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................AAGAAGTGCTCACGTCAGGTCTCTTCC.................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGTGCTCCCGTTCCTAAGAAGTG....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TGTGCTCCTGATGTCTGACTGGC................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GCTACAACCTGAGCCCTCAGTTCACGC........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGATGTCTGACTGGCTCCCCAAGA........................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCCTTGATAGTGAAGAGATGGGAGGCAGGGCACACCCAAAGAAATCTCCTGTGCTCCTGATGTCTGACTGGCTCCCCAAGACTTCTCTGTCCTCATGGTTCTGAGCTGATCCCTCAGGACCCCTGATCTGCTGTGATCTAGTGCTCCCGTTCCTAAGAAGTGCTCACGTCAGGTCTCTTCCTGTGCCCTTCCCCTCCAGCGCTCTCCCAGATGGGCTACAACCTGAGCCCTCAGTTCACGCAGCTCCTG .....................................................................................................................(((.....((((((.(((((....(((((.............)))))))))).))))))...))).................................................................... ...........................................................................................................108.......................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................AGACTTCTCTGTCCTCgc........................................................................................................................................................... | 18 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |