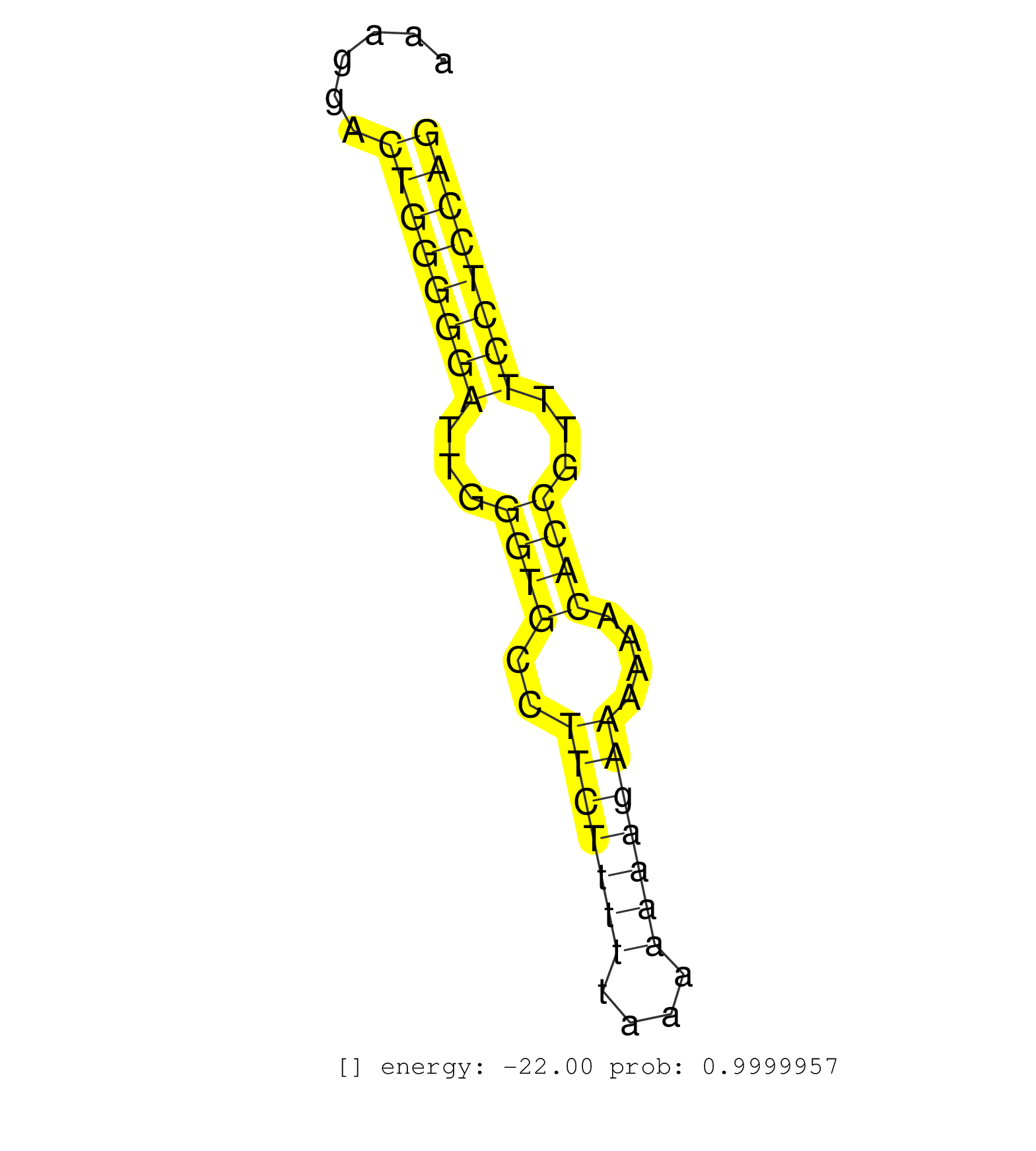
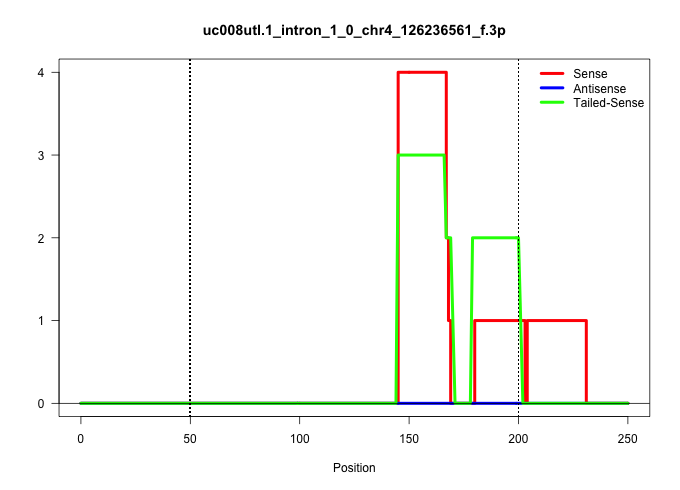
| Gene: Clspn | ID: uc008utl.1_intron_1_0_chr4_126236561_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(9) TESTES |
| TTTTGCCACAGAAATTTCTTTTAAATATATTTTTAGATATGCTTTGCTTCAATGAGTTCTTACTGGCAGTTGTGTAATCCCATAAGGTGATATATCACATGTGTAATTAAATATGTTTTTAAGTAGCTTTGAAGAAGGACAAAGGACTGGGGGATTGGGTGCCTTCTTTTTAAAAAAAGAAAAAACACCGTTTCCTCCAGATTCTGATGAAGAGATTTTTGTGAGCAAGAAACCTAAGAGCAGGAAAGTG ..................................................................................................................................................((((((((...((((..(((((((....)))))))....))))...)))))))).................................................. ............................................................................................................................................141........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTCT................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................AAAAAACACCGTTTCCTCCAGt................................................. | 22 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTCTTaaa............................................................................... | 26 | aaa | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTCTTT................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTCTaaa................................................................................ | 25 | aaa | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTaa................................................................................... | 22 | aa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................ACTGGGGGATTGGGTGCCTTCTT.................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................AAAAACACCGTTTCCTCCAGATT............................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGATGAAGAGATTTTTGTGAGCAAGAA................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................AAAAAACACCGTTTCCTCCAGtt................................................ | 23 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| TTTTGCCACAGAAATTTCTTTTAAATATATTTTTAGATATGCTTTGCTTCAATGAGTTCTTACTGGCAGTTGTGTAATCCCATAAGGTGATATATCACATGTGTAATTAAATATGTTTTTAAGTAGCTTTGAAGAAGGACAAAGGACTGGGGGATTGGGTGCCTTCTTTTTAAAAAAAGAAAAAACACCGTTTCCTCCAGATTCTGATGAAGAGATTTTTGTGAGCAAGAAACCTAAGAGCAGGAAAGTG ..................................................................................................................................................((((((((...((((..(((((((....)))))))....))))...)))))))).................................................. ............................................................................................................................................141........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|