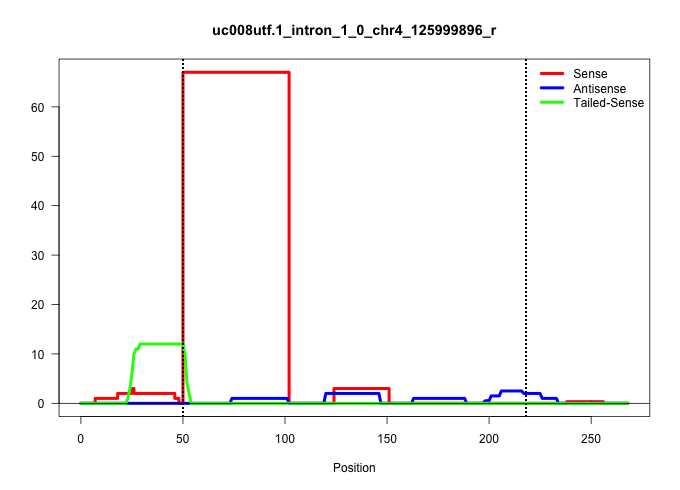
| Gene: Tekt2 | ID: uc008utf.1_intron_1_0_chr4_125999896_r | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(2) PIWI.ip |
(1) TDRD1.ip |
(18) TESTES |
| GGCTGGAGTCCAGAACCTACCGGTCCAATGTGGAGCTCTGCCGGGACCAGGTACAAGGGGCCCCGGTGGGGACCACCCCTGTCTGCTAAGGAGACCTCAGTATTACCCCTCTTCGCACGTTTCTTGTGGAACAAGCATTCTGGTCCAAGGCAGTTATGCGACCAGCTGAAGGGTCTTGCGGCTGAACTGAAAACCCCATCCTGCCCCTCTGTCTTCAGACACAGTATGGCCTCATTGATGAAGTCCACCAGTTAGAGGCAACCATCAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTACAAGGGGCCCCGGTGGGGACCACCCCTGTCTGCTAAGGAGACCTCAGTA...................................................................................................................................................................... | 52 | 1 | 67.00 | 67.00 | 67.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGTGGAACAAGCATTCTGGTCCAAGGC..................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................AATGTGGAGCTCTGCCGGGACCAGac........................................................................................................................................................................................................................ | 26 | ac | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................AATGTGGAGCTCTGCCGGGACCAGacac...................................................................................................................................................................................................................... | 28 | acac | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................CCAATGTGGAGCTCTGCCGGGACCAGac........................................................................................................................................................................................................................ | 28 | ac | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAATGTGGAGCTCTGCCGGGACCAGaca....................................................................................................................................................................................................................... | 28 | aca | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................ATGTGGAGCTCTGCCGGGACCAGa......................................................................................................................................................................................................................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................CAATGTGGAGCTCTGCCGGGACCAGac........................................................................................................................................................................................................................ | 27 | ac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAATGTGGAGCTCTGCCGGGACC............................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCCAATGTGGAGCTCTGCCGGGACCAGac........................................................................................................................................................................................................................ | 29 | ac | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ACCGGTCCAATGTGGAGCTCTGCCGGGA.............................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......GTCCAGAACCTACCGGTCC.................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................GTGGAGCTCTGCCGGGACCAGa......................................................................................................................................................................................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................TGAAGTCCACCAGTTAGA............ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GGCTGGAGTCCAGAACCTACCGGTCCAATGTGGAGCTCTGCCGGGACCAGGTACAAGGGGCCCCGGTGGGGACCACCCCTGTCTGCTAAGGAGACCTCAGTATTACCCCTCTTCGCACGTTTCTTGTGGAACAAGCATTCTGGTCCAAGGCAGTTATGCGACCAGCTGAAGGGTCTTGCGGCTGAACTGAAAACCCCATCCTGCCCCTCTGTCTTCAGACACAGTATGGCCTCATTGATGAAGTCCACCAGTTAGAGGCAACCATCAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TTCTTGTGGAACAAGCATTCTGGTCCA......................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................ACCCCTGTCTGCTAAGGAGACCTCAGTA...................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCCCTCTGTCTTCAGACACAGTA.......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................TGAAGTCCACCAGTgagg................ | 18 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................CCCTCTTCGCACGtaag..................................................................................................................................................... | 17 | taag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................CTCTGTCTTCAGACACAGTATGGCCTCA.................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGCTGAAGGGTCTTGCGGCTGAACTG............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TCCTGCCCCTCTGTCTTCA................................................... | 19 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |