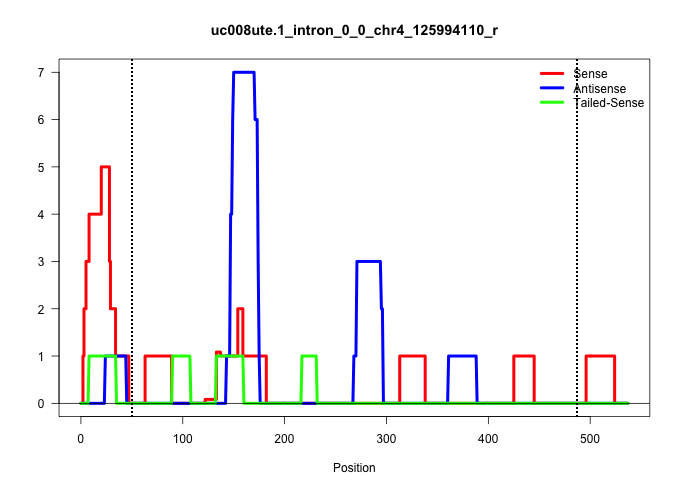
| Gene: Adprhl2 | ID: uc008ute.1_intron_0_0_chr4_125994110_r | SPECIES: mm9 |
 |
 |
|
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(14) TESTES |
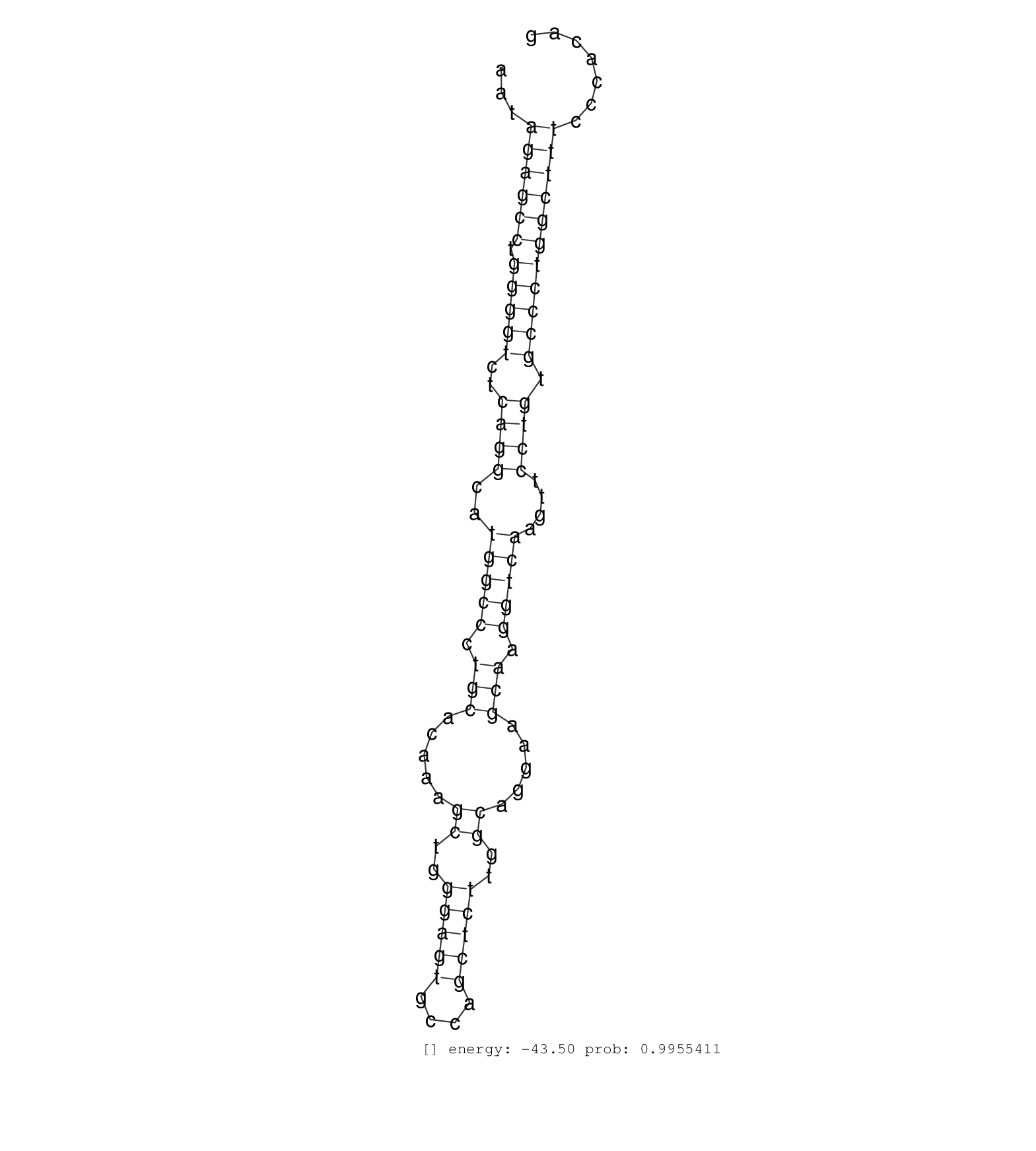
| GCTGCTGGACCAGGACGTGGTGAGCCGAGAGGAAGTGGTGTCCGAGCTAGGTGAGTGGACCCCTGCTGACCGTGGAGCAGAGAAGGCTTCTGGGACAGTAGAGCTCCTCTTTGTAGGGTCCCTAGCTGGGCTTTGTACACTGTGCCTGCAGCCTTGACGATCAGTATCCCCAGAAAAGGCAGCTGGCAGCCAGTTTGTTCTCTAGTGTGGTTGTGGATGTAAATTTGCCTGGTACCTGTGCCCTGGGCACTGTGTGGAGGCCTTCACAGTGACCCGCCTTTCAACAGCCCTGGGAGAGAGAGAGAGCAAGGATTAAAGACGAGCGCAGGACTAAGTGGGACCCTTGCCTGGCCACCAGCAGGCTTGTTGGGTCCCGAGTAGTCTTACAATAGAGCCTGGGGTCTCAGGCATGGCCCTGCACAAAGCTGGGAGTGCCAGCTCTTGGCAGGAAGCAAGGTCAAGTTCCTGTGCCCTGGCTTTCCCACAGGGAATGGCATTGCCGCCTTTGAATCTGTGCCCACCGCCATCTACTGCTTC ......................................................................................................................................................................................................................................................................................................................................................................................................((((((.(((((..((((..(((((.(((.....((..(((((....)))))..)).....))).)))))....)))).)))))))))))......................................................... ...................................................................................................................................................................................................................................................................................................................................................................................................388................................................................................................487................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................................TAAAGACGAGCGCAGGACTAAGTGG....................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGCTGACCGTGGAGCAGAGAAGGCTT................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................TGACGATCAGTATCCCCAGAAAAGGCAG................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................TGTACACTGTGCCTGCAGCCTTGACGt......................................................................................................................................................................................................................................................................................................................................................................................... | 27 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTACACTGTGCCTGCAGCCTTGACG.......................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........ACCAGGACGTGGTGAGCCGAGAGGAAt...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..TGCTGGACCAGGACGTGGTGAGCCGA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGGGACAGTAGAGCatct............................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | atct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................CTGGGAGTGCCAGCTCTTGG............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................TGAGCCGAGAGGAAGTGGTGTCCGAGC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTGCCGCCTTTGAATCTGTGCCCACCGC............. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ACCAGGACGTGGTGAGCCGAGAGGAA....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...GCTGGACCAGGACGTGGTGAGCCGA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....TGGACCAGGACGTGGTGAGCCGAG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGCTGGGCTTTGTA................................................................................................................................................................................................................................................................................................................................................................................................................ | 15 | 12 | 0.08 | 0.08 | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - |
| GCTGCTGGACCAGGACGTGGTGAGCCGAGAGGAAGTGGTGTCCGAGCTAGGTGAGTGGACCCCTGCTGACCGTGGAGCAGAGAAGGCTTCTGGGACAGTAGAGCTCCTCTTTGTAGGGTCCCTAGCTGGGCTTTGTACACTGTGCCTGCAGCCTTGACGATCAGTATCCCCAGAAAAGGCAGCTGGCAGCCAGTTTGTTCTCTAGTGTGGTTGTGGATGTAAATTTGCCTGGTACCTGTGCCCTGGGCACTGTGTGGAGGCCTTCACAGTGACCCGCCTTTCAACAGCCCTGGGAGAGAGAGAGAGCAAGGATTAAAGACGAGCGCAGGACTAAGTGGGACCCTTGCCTGGCCACCAGCAGGCTTGTTGGGTCCCGAGTAGTCTTACAATAGAGCCTGGGGTCTCAGGCATGGCCCTGCACAAAGCTGGGAGTGCCAGCTCTTGGCAGGAAGCAAGGTCAAGTTCCTGTGCCCTGGCTTTCCCACAGGGAATGGCATTGCCGCCTTTGAATCTGTGCCCACCGCCATCTACTGCTTC ......................................................................................................................................................................................................................................................................................................................................................................................................((((((.(((((..((((..(((((.(((.....((..(((((....)))))..)).....))).)))))....)))).)))))))))))......................................................... ...................................................................................................................................................................................................................................................................................................................................................................................................388................................................................................................487................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................GCAGCCTTGACGATCAGTATCCCCAGA........................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CCTGCAGCCTTGACGATCAGTATCCCCA............................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................ACCCGCCTTTCAACAGCCCTGGGAGA................................................................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGCCTTGACGATCAGTATCCCCAGAA.......................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................GCCTTGACGATCAGTATCCCCAGAA.......................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................GTGACCCGCCTTTCAACAGCCCTGGGA.................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGCCTTGACGATCAGTATCCCCAGAAA......................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................GCCTGCAGCCTTGACGATCAGTATCCCC.............................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CCGAGAGGAAGTGGTGTCCGA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................GCTTGTTGGGTCCCGAGTAGTCTTACAA.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |