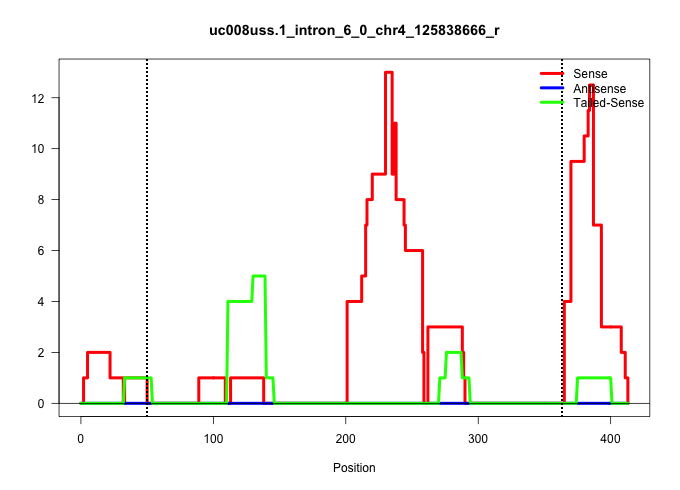
| Gene: 1700029G01Rik | ID: uc008uss.1_intron_6_0_chr4_125838666_r | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| ATAGTTGAGCAACTTGACATATGACAGCCCTCCAGACTACCTGCGGACAGGTGAGCCCTCCCTCGGTACCAGATCTATGTGGGAGGTGAACTGGCTAGGTTGTTGGAGGGAGTCACTGGCTGCGGAGGGAAGGGCTACACACTAGAGTGACCTTCTTTTAGTCTCCTGCCCTGAGACCTGCAGGGTCCTATTTGACTACCAACCCGAGGCCCCAGATGAGCTGGCACTGCAGAAGGGGGACCTGGTGAAAGTTCTGAGAAAGGTATGAGAGGATTAGGAGTTACAGAAGAGTAAGGAGTGGGGCCCAGGCAAGAAGAACTTGAGCGATGCTCACCTCCCCCCACTGGCTTGTTTCCTAACCAGACAACTGAGGATAAGGGCTGGTGGGAAGGAGAATGCCAAGGCAGAAGAGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................................................................................................................AGGATAAGGGCTGGTGG.......................... | 17 | 2 | 5.50 | 5.50 | 5.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................ACCCGAGGCCCCAGATGAGCTGGCACTGCAGAAG.................................................................................................................................................................................. | 34 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTCACTGGCTGCGGAGGGAAGGGCTACAt................................................................................................................................................................................................................................................................................. | 29 | t | 4.00 | 0.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................AACTGAGGATAAGGGCTGGTGGGAAGGA.................... | 28 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................AGAAGGGGGACCTGGTGAAAGTTCTGAG........................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................ATGAGCTGGCACTGCAGAAGGGG............................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................GGACCTGGTGAAAGTTCTGAGA.......................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGACTACCTGCGGACAG........................................................................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................AGGGCTACACACgtgc........................................................................................................................................................................................................................................................................... | 16 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..AGTTGAGCAACTTGACATAT....................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGCTGGCACTGCAGAAGGGGGACCTG......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................GTGGGAAGGAGAATGCCAAGGCAGAAGAGG | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGACTACCTGCGGACAGtctc....................................................................................................................................................................................................................................................................................................................................................................... | 21 | tctc | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................GTATGAGAGGATTAGGAGTTACAGAA............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................TGGGAAGGAGAATGCCAAGGCAGAAGA.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....TGAGCAACTTGACATATGACAGCCCTC............................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................AAGGGCTGGTGGGAAGGAGAATGCCt............ | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CAGATGAGCTGGCACTGCAGAAGGGG............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................GTATGAGAGGATTAGGAGTTACAGAAGA........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................ACTGGCTAGGTTGTTGGAGG................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................CTGGTGGGAAGGAGAATGCCAAGGCAGA..... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................GTATGAGAGGATTAGGAGTTACAGAAG............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CTGGCACTGCAGAAGGGGGACCTGG........................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................................................................................GGAGTTACAGAAGAtgaa....................................................................................................................... | 18 | tgaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................CACTGGCTGCGGAGGGAAGGGCTAC................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................................................................................GATTAGGAGTTACgggg............................................................................................................................. | 17 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ATAGTTGAGCAACTTGACATATGACAGCCCTCCAGACTACCTGCGGACAGGTGAGCCCTCCCTCGGTACCAGATCTATGTGGGAGGTGAACTGGCTAGGTTGTTGGAGGGAGTCACTGGCTGCGGAGGGAAGGGCTACACACTAGAGTGACCTTCTTTTAGTCTCCTGCCCTGAGACCTGCAGGGTCCTATTTGACTACCAACCCGAGGCCCCAGATGAGCTGGCACTGCAGAAGGGGGACCTGGTGAAAGTTCTGAGAAAGGTATGAGAGGATTAGGAGTTACAGAAGAGTAAGGAGTGGGGCCCAGGCAAGAAGAACTTGAGCGATGCTCACCTCCCCCCACTGGCTTGTTTCCTAACCAGACAACTGAGGATAAGGGCTGGTGGGAAGGAGAATGCCAAGGCAGAAGAGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|