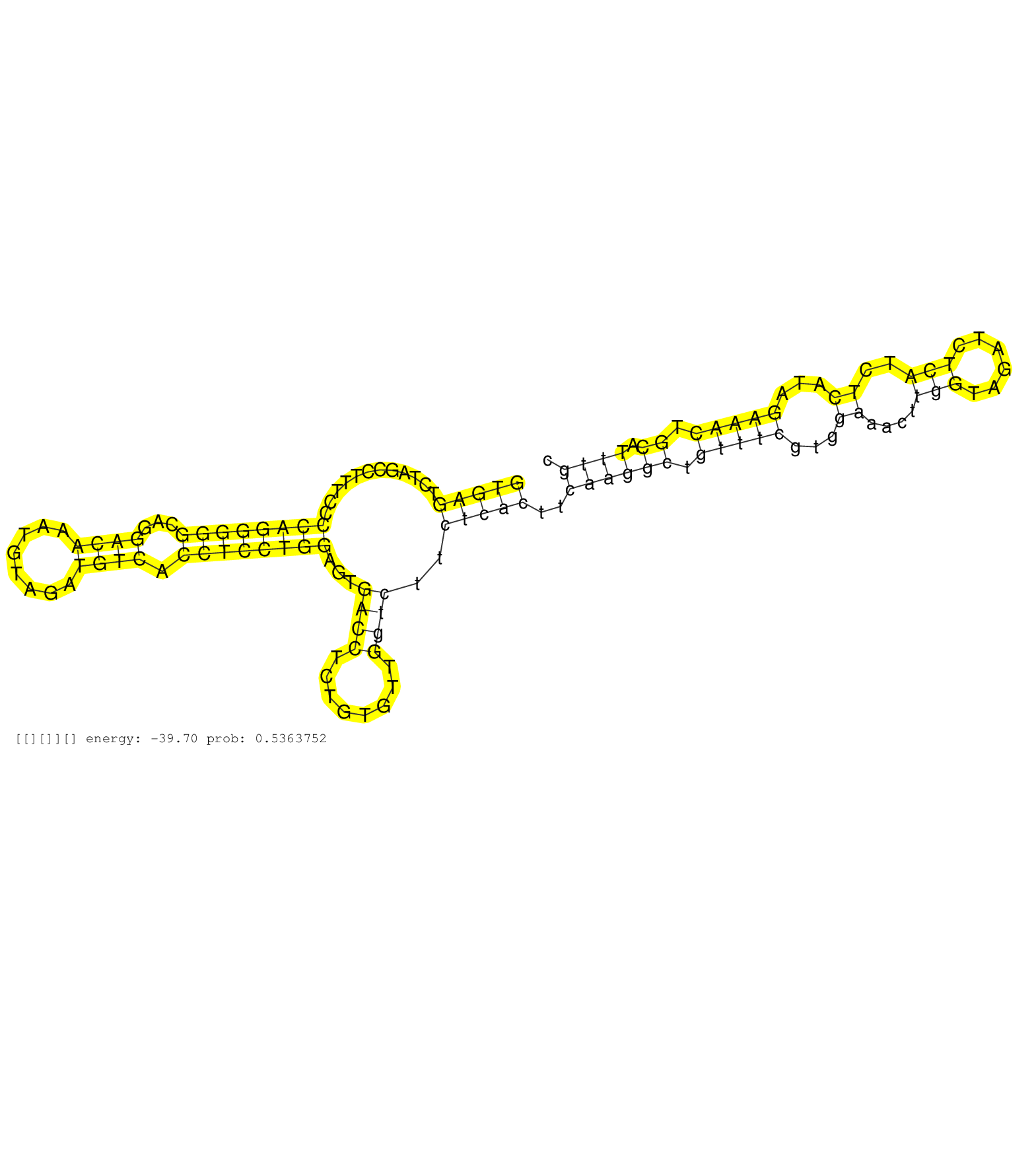

| Gene: Cap1 | ID: uc008uoj.1_intron_7_0_chr4_122542519_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(9) TESTES |
| GACGCGGCCATGTTTTACACAAATCGTGTCCTCAAGGAGTACAGAGATGTGTGAGTCTAGCCTTTCCCCAGGGGGCAGGACAAATGTAGATGTCACCTCCTGGAGTGACCTCTGTGTTGGTCTTCTCACTTCAAGGCTGTTTCGTGGAAACTTGGTAGATCTCATCTCATAGAAACTGCATTTGCAAGCAGGAAATATTTCTGCTTCCATTTCTTTTTTTTTTTTTTTTTAAAGATTTATTTATTTATTA ..................................................(((((............((((((((...((((........)))).))))))))...((((........))))..)))))..((((((.(((((...((....(((......)))..))...))))).)).)))).................................................................. ..................................................51....................................................................................................................................185............................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTCTAGCCTTTCCCCAGGGGGCAGGACAAATGTAGATGTCACCTCCTG.................................................................................................................................................... | 52 | 1 | 64.00 | 64.00 | 64.00 | - | - | - | - | - | - | - | - |
| ...................CAAATCGTGTCCTCAAGGAGTACAGA............................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................GTAGATCTCATCTCATAGAAACTGCAT..................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................GGTAGATCTCATCTCATAGAA............................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................CAAATGTAGATGTCACCTCCT..................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ....................AAATCGTGTCCTCAAGGAGT.................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..........................TGTCCTCAAGGAGTACAGAGATGTGg...................................................................................................................................................................................................... | 26 | g | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - |
| ......................ATCGTGTCCTCAAGGAGTACAGAGATGTGgat.................................................................................................................................................................................................... | 32 | gat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGGTAGATCTCATCTCATAGAAACTGC....................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ............................TCCTCAAGGAGTACAGAGATGTGgat.................................................................................................................................................................................................... | 26 | gat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| ................ACACAAATCGTGTCCTCAAGGAGTACAG.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| GACGCGGCCATGTTTTACACAAATCGTGTCCTCAAGGAGTACAGAGATGTGTGAGTCTAGCCTTTCCCCAGGGGGCAGGACAAATGTAGATGTCACCTCCTGGAGTGACCTCTGTGTTGGTCTTCTCACTTCAAGGCTGTTTCGTGGAAACTTGGTAGATCTCATCTCATAGAAACTGCATTTGCAAGCAGGAAATATTTCTGCTTCCATTTCTTTTTTTTTTTTTTTTTAAAGATTTATTTATTTATTA ..................................................(((((............((((((((...((((........)))).))))))))...((((........))))..)))))..((((((.(((((...((....(((......)))..))...))))).)).)))).................................................................. ..................................................51....................................................................................................................................185............................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|