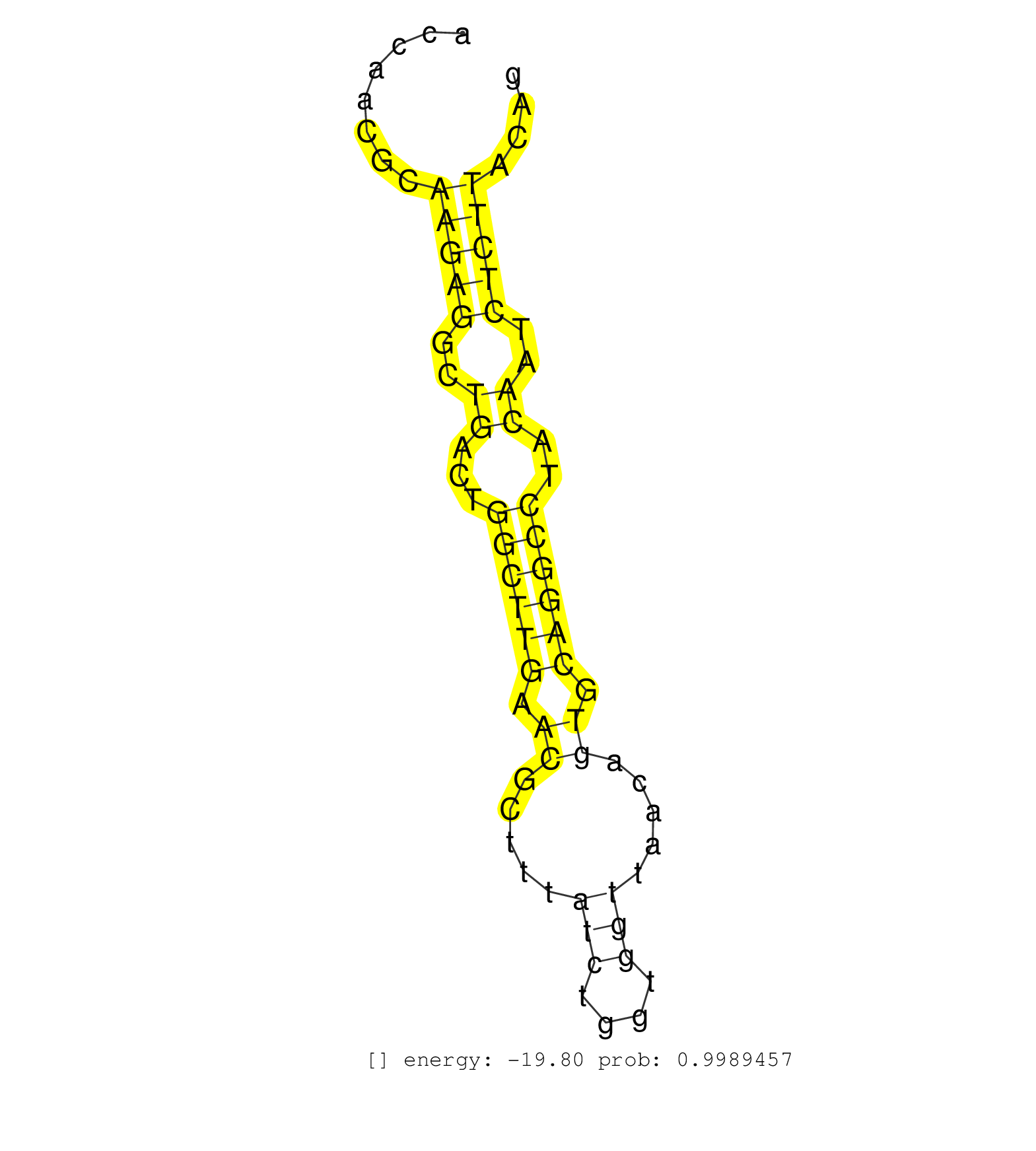
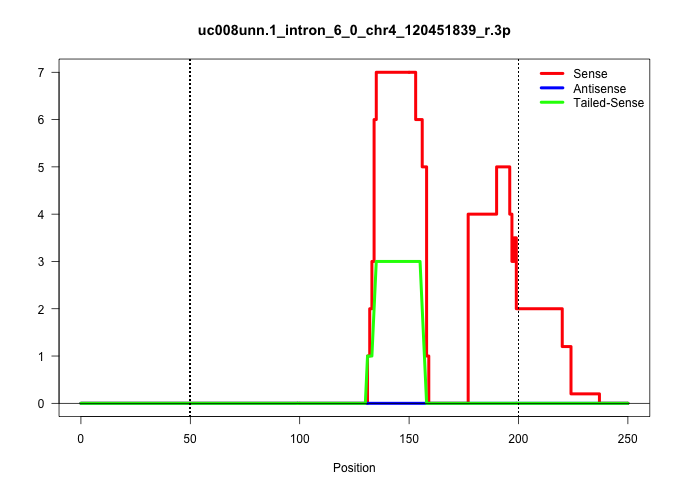
| Gene: Nfyc | ID: uc008unn.1_intron_6_0_chr4_120451839_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| AAGAGTAAACACCATGCCTTGACCTGGCAGTGACAGGTGCTATGTCTTACCCTACTCGGGAATTTATTTTACTCTGGACTCTCGAGAGTCTACCGCTTTATTTTGTAGTTCTTATCTAAAGGTTGGCACCAACGCAAGAGGCTGACTGGCTTGAACGCTTTATCTGGTGGTTAACAGTGCAGGCCTACAATCTCTTACAGATGATCAGTGCAGAAGCCCCTGTGCTGTTTGCTAAGGCAGCCCAGATTTT .......................................................................................................................................(((((..((...((((((.((.....(((....))).....)).))))))..))..)))))...................................................... ...............................................................................................................................128.....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................CAAGAGGCTGACTGGCTTGAAC.............................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCAGGCCTACAATCTCTTACA................................................... | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CGCAAGAGGCTGACTGGCTTGAACGC............................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CAAGAGGCTGACTGGCTTGAACa............................................................................................. | 23 | a | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCAGGCCTACAATCTCTT...................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCAGGCCTACAATCTCTTA..................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CAAGAGGCTGACTGGCTTGAACGC............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................GCAAGAGGCTGACTGGCTTGAACGC............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................TCTCTTACAGATGATCAGTGCAGAAGCCCC.............................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................ACGCAAGAGGCTGACTGGCTTGAAagc............................................................................................ | 27 | agc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CAAGAGGCTGACTGGCTTGAACGCT........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGAGGCTGACTGGCTTGAAa.............................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCAGGCCTACAATCTCTTACAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGAGGCTGACTGGCTTGAACGCT........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGAGGCTGACTGGCTTGAACGt............................................................................................ | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................AAGAGGCTGACTGGCTTGAACGC............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................ACGCAAGAGGCTGACTGGCTTG................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGATCAGTGCAGAAGCCCCTGTG.......................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................................................................................................................AGATGATCAGTGCAGAAGCCCCTGTG.......................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...................................................................................................................................................................................................................AGAAGCCCCTGTGCTGTTTGCTAA............... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ............................................................................................................................................................................................................................TGTGCTGTTTGCTAAGG............. | 17 | 5 | 0.20 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGAGTAAACACCATGCCTTGACCTGGCAGTGACAGGTGCTATGTCTTACCCTACTCGGGAATTTATTTTACTCTGGACTCTCGAGAGTCTACCGCTTTATTTTGTAGTTCTTATCTAAAGGTTGGCACCAACGCAAGAGGCTGACTGGCTTGAACGCTTTATCTGGTGGTTAACAGTGCAGGCCTACAATCTCTTACAGATGATCAGTGCAGAAGCCCCTGTGCTGTTTGCTAAGGCAGCCCAGATTTT .......................................................................................................................................(((((..((...((((((.((.....(((....))).....)).))))))..))..)))))...................................................... ...............................................................................................................................128.....................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|