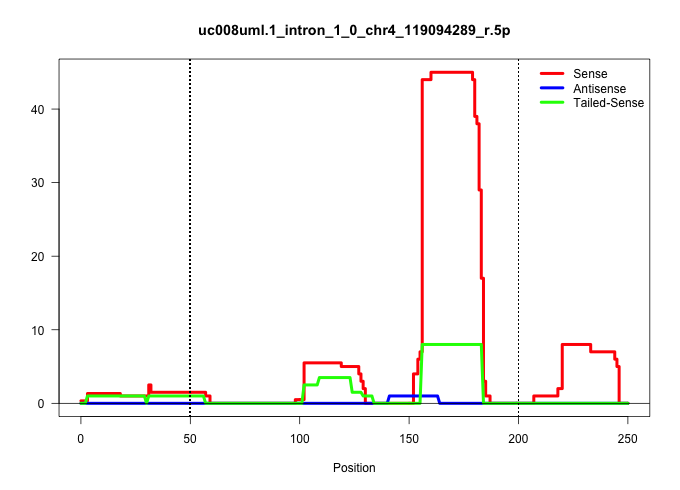
| Gene: Ppcs | ID: uc008uml.1_intron_1_0_chr4_119094289_r.5p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(23) TESTES |
| ACCTGGTGGCTGAGTTGCCGCGTCCTGCGGGTGCTGCGCGCTGGGCCGAGGTTATGGCTCGCTTTGCGGCCAGGCTGGGCGAGCAGGGCCGGCGGGTGGTGCTGATCACATCTGGAGGCACCAAGGTCCCCCTGGAAGCGCGAGCCGTGCGCTTTCTGGACAACTTCAGTAGCGGGCGACGCGGAGCCGCGTCGGCGGAGGTCTTCCTGGCTGCCGGCTATGGAGTCCTGTTCTTGTACCGAGCGCGCTC .................................................................................................((((((..(((....))).))))))...((((....(((((((((.....))))))))).))))..((((....(((.....)))))))................................................................ .................................................................................................98........................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGCG................................................................... | 27 | 1 | 28.00 | 28.00 | 11.00 | 5.00 | 5.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGC.................................................................... | 26 | 1 | 24.00 | 24.00 | 4.00 | 1.00 | 10.00 | - | - | 1.00 | - | - | - | 4.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGCGG.................................................................. | 28 | 1 | 24.00 | 24.00 | 6.00 | 5.00 | - | 1.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGAGTCCTGTTCTTGTACCGAGCGC.... | 26 | 1 | 9.00 | 9.00 | - | 1.00 | - | 4.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................TATGGAGTCCTGTTCTTGTACCGAGC...... | 26 | 1 | 8.00 | 8.00 | - | - | 1.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACG..................................................................... | 25 | 1 | 7.00 | 7.00 | - | 1.00 | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGCGt.................................................................. | 28 | t | 6.00 | 28.00 | - | 2.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCTGGACAACTTCAGTAGCGGGCGACG..................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TTTCTGGACAACTTCAGTAGCGGGCGAC...................................................................... | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGCGGGTGCTGCGCGCTGGGCCGAGG....................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGtgg.................................................................. | 28 | tgg | 2.00 | 7.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAGGTCCC........................................................................................................................ | 28 | 2 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCTGGACAACTTCAGTAGCGGGCGA....................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCAtcaa.............................................................................................................................. | 22 | tcaa | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGCGGA................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCTGGACAACTTCAGTAGCGGGCGAC...................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAGGTC.......................................................................................................................... | 26 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ................................................................................................................................................................CAACTTCAGTAGCGGGCGACGCGGAGC............................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGGCTGCCGGCTATGGAGTCCTGTTC................. | 26 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CTGGACAACTTCAGTAGCGGGCGACGC.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TTTCTGGACAACTTCAGTAGCGGGCGA....................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGGACAACTTCAGTAGCGGGCGACGg.................................................................... | 26 | g | 1.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................ATCTGGAGGCACCAAGGTCCCCCTt.................................................................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAGGTCC......................................................................................................................... | 27 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGAGTCCTGTTCTTGTACCGAGCG..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGCTGCGCGCTGGGCCGAGGTTATGt................................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGAGTCCTGTTCTTGTACCGAGC...... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGCTGCGCGCTGGGCCGAGGTTATGGCT............................................................................................................................................................................................... | 28 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAGGT........................................................................................................................... | 25 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGGTGGCTGAGTTGCCGCGTCCTGCGGGT.......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGGTGGCTGAGTTGCCGCGTCCTGCta............................................................................................................................................................................................................................ | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAG............................................................................................................................. | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ..................................................................................................GTGCTGATCACATCTGGAGGC................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................TGATCACATCTGGAGGCACCAAGGTCt......................................................................................................................... | 27 | t | 0.50 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGCTGCGCGCTGGGCCGAGGTTAT................................................................................................................................................................................................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ...............................TGCTGCGCGCTGGGCCGAGGTTATGG................................................................................................................................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ACCTGGTGGCTGAGTTGCCGCGTCCTGCGGGTGCTGCGCGCTGGGCCGAGGTTATGGCTCGCTTTGCGGCCAGGCTGGGCGAGCAGGGCCGGCGGGTGGTGCTGATCACATCTGGAGGCACCAAGGTCCCCCTGGAAGCGCGAGCCGTGCGCTTTCTGGACAACTTCAGTAGCGGGCGACGCGGAGCCGCGTCGGCGGAGGTCTTCCTGGCTGCCGGCTATGGAGTCCTGTTCTTGTACCGAGCGCGCTC .................................................................................................((((((..(((....))).))))))...((((....(((((((((.....))))))))).))))..((((....(((.....)))))))................................................................ .................................................................................................98........................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................GCCGGCTATGGAGTCCTGTTCTTGTA............ | 26 | 1 | 5.00 | 5.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGCCGTGCGCTTTCTGGACAAC...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |