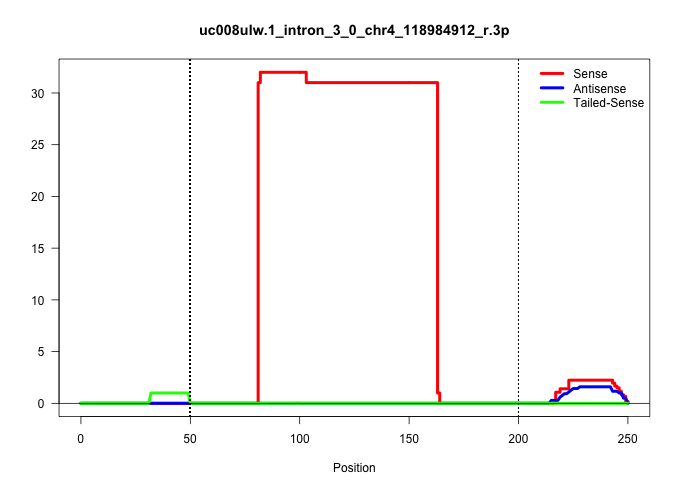
| Gene: Ppih | ID: uc008ulw.1_intron_3_0_chr4_118984912_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(17) TESTES |
| TACACTAAGACAACTGTGCATTGATAAAAGAATTTAGATTCTCAGGGGTGGGGACTCTGCATCCTATTCATCCAATGTCCTTGTCTAGGCCTGGGCTCATAATAGGTGCTCGGTTAGGTTGTAGTTAGAGCTTCAGACTTTGTGGGAAATCACCTACAGTGGCCAGAGAAACTTATGTAGGTGTATTTTGTTTCTGTTAGGCAAACAGTGGTCCCAGTACAAATGGCTGCCAGTTCTTTATCACGTGTTC .....................................................................................................((((((.((((((((.(((..(((....)))...)))..((((((......)))))).))))).)))..)))))).......................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................TGTCTAGGCCTGGGCTCATAATAGGTGCTCGGTTAGGTTGTAGTTAGAGCTT..................................................................................................................... | 52 | 1 | 32.00 | 32.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTTAGATTCTCAGGactt........................................................................................................................................................................................................ | 18 | actt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GTCTAGGCCTGGGCTCATAAT................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCTGCCAGTTCTTTATCACGTGTT. | 26 | 6 | 0.50 | 0.50 | - | - | - | - | - | 0.33 | - | - | 0.17 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TACAAATGGCTGCCAGTTCTTTATCACGT.... | 29 | 6 | 0.33 | 0.33 | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | 0.17 | - | - | - | - |
| .........................................................................................................................................................................................................................TACAAATGGCTGCCAGTTCTTTATCAC...... | 27 | 7 | 0.29 | 0.29 | - | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TACAAATGGCTGCCAGTTCTTTATCA....... | 26 | 7 | 0.29 | 0.29 | - | - | - | - | - | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TACAAATGGCTGCCAGTTCTTTATCACG..... | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCTGCCAGTTCTTTATCACGTGT.. | 25 | 6 | 0.17 | 0.17 | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCTGCCAGTTCTTTATCACGTG... | 24 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCTGCCAGTTCTTTATCACGTGTTC | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CAAATGGCTGCCAGTTCTTTATCACGTGT.. | 29 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CAAATGGCTGCCAGTTCTTTATCACGTG... | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| ........................................................................................................................................................................................................................GTACAAATGGCTGCCAGTTCTTTATC........ | 26 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - |
| TACACTAAGACAACTGTGCATTGATAAAAGAATTTAGATTCTCAGGGGTGGGGACTCTGCATCCTATTCATCCAATGTCCTTGTCTAGGCCTGGGCTCATAATAGGTGCTCGGTTAGGTTGTAGTTAGAGCTTCAGACTTTGTGGGAAATCACCTACAGTGGCCAGAGAAACTTATGTAGGTGTATTTTGTTTCTGTTAGGCAAACAGTGGTCCCAGTACAAATGGCTGCCAGTTCTTTATCACGTGTTC .....................................................................................................((((((.((((((((.(((..(((....)))...)))..((((((......)))))).))))).)))..)))))).......................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................CTGCCAGTTCTTTATCACGTGTTCacat | 28 | acat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GGCTGCCAGTTCTTTATCACGTGTttct.. | 28 | ttct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AATGGCTGCCAGTTCTTTATCACGTGT.. | 27 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | - | - | - | - | - |
| .......................................................................................................................................................................................................................AGTACAAATGGCTGCCAGTTCTTTATCA....... | 28 | 7 | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.14 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGCTGCCAGTTCTTTATCACGTGTTC | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AAATGGCTGCCAGTTCTTTATCACGTGTa.. | 29 | a | 0.17 | 0.00 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................GCCAGTTCTTTATCACGT.... | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CAAATGGCTGCCAGTTCTTTATCACGTGT.. | 29 | 6 | 0.17 | 0.17 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GGCTGCCAGTTCTTTATCACGTGTTC | 26 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GCTGCCAGTTCTTTATCACGTGTTC | 25 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| ............................................................................................................................................................................................................................AAATGGCTGCCAGTTCTTTATCACGTGT.. | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CAAATGGCTGCCAGTTCTTTATCA....... | 24 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| ..........................................................................................................................................................................................................................ACAAATGGCTGCCAGTTCTTTATCA....... | 25 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |