
| Gene: Cdc20 | ID: uc008ukc.1_intron_8_0_chr4_118109448_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(17) TESTES |
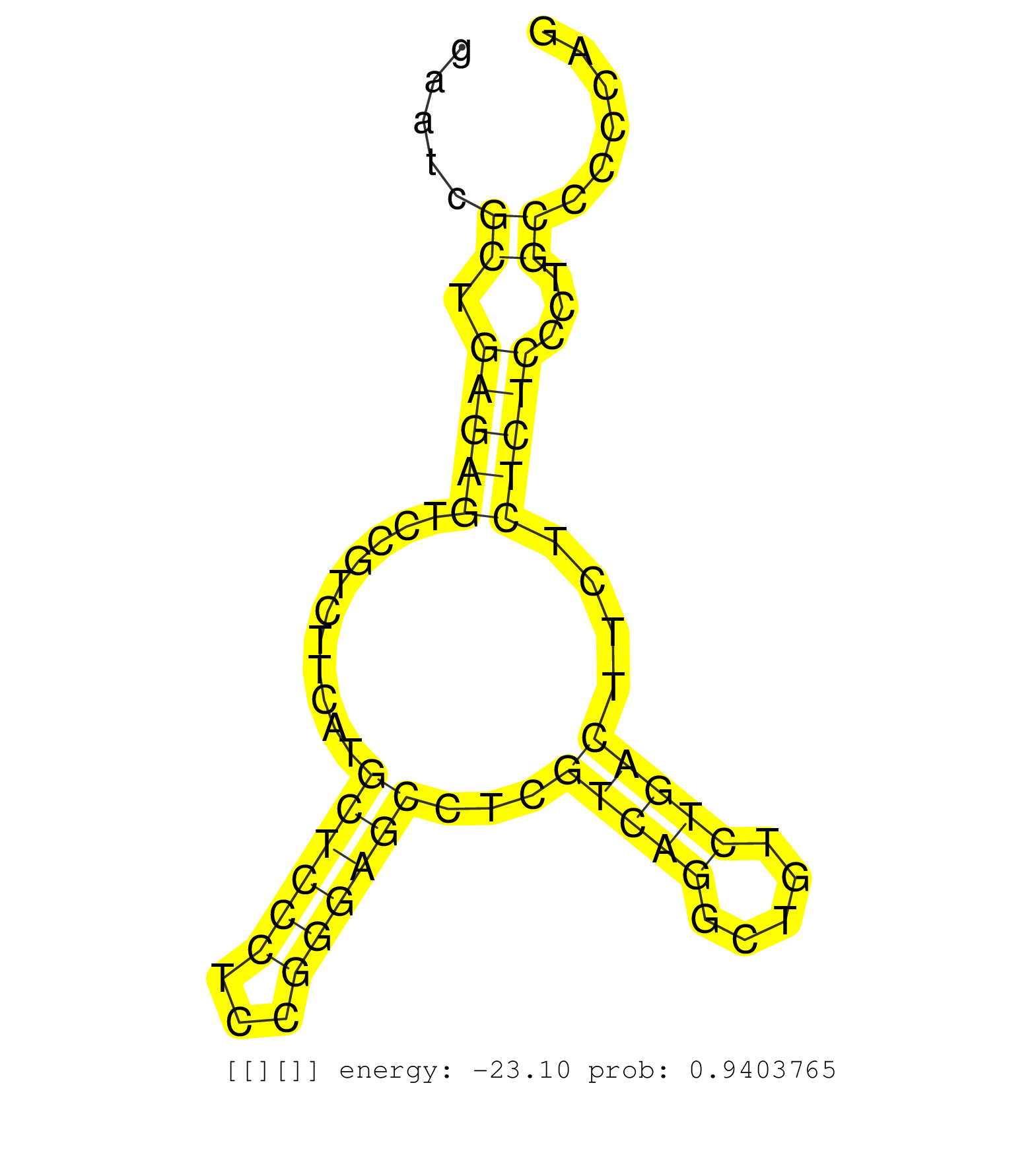
| GGCCGCCAACAGATCACACAGCGCCGGGCGGACCCCGGGCCGAACTCCTGGTGAGTGGGTGGAGGGAGCTGGAATCGCTGAGAGTCCGTCTTCATGCTCCCTCCGGGAGCCTCGTCAGGCTGTCTGACTTCTCTCTCCCTGCCCCCAGGCAAATCTAGTTCCAAGGTTCAGACCACCCCTAGCAAACCTGGAGGTGAC ............................................................................((.(((((...........((((((...))))))...(((((.....)))))....)))))...))........................................................ .......................................................................72..........................................................................148................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................GCTGAGAGTCCGTCTTCATGCTCCCTCCGGGAGCCTCGTCAGGCTGTCTGAC...................................................................... | 52 | 1 | 90.00 | 90.00 | 90.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CAACAGATCACACAGCGCCGGGCGta...................................................................................................................................................................... | 26 | ta | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ......CAACAGATCACACAGCGCCGGGCGGAC..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ACAGATCACACAGCGCCG............................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GCTGAGAGTCCGTCTTCAT....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TAGTTCCAAGGTTCAGACCACCCC................... | 24 | 2 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TAGTTCCAAGGTTCAGACCACCCCTAa................ | 27 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CTTCTCTCTCCCTGCCCCCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CAACAGATCACACAGCGCCGGGCGGACC.................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ACAGATCACACAGCGCCGGGCGGACC.................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................TGGGTGGAGGGAGCTGGAATCGa........................................................................................................................ | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CAAATCTAGTTCCAAGGTTCAGACCACCC.................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ......................................................................................................................................................AAATCTAGTTCCAAGGTTCAGACCACCC.................... | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................................................................................................................TAGTTCCAAGGTTCAGACCACCCCTAGC............... | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| GGCCGCCAACAGATCACACAGCGCCGGGCGGACCCCGGGCCGAACTCCTGGTGAGTGGGTGGAGGGAGCTGGAATCGCTGAGAGTCCGTCTTCATGCTCCCTCCGGGAGCCTCGTCAGGCTGTCTGACTTCTCTCTCCCTGCCCCCAGGCAAATCTAGTTCCAAGGTTCAGACCACCCCTAGCAAACCTGGAGGTGAC ............................................................................((.(((((...........((((((...))))))...(((((.....)))))....)))))...))........................................................ .......................................................................72..........................................................................148................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................GCTGGAATCGCTGAGAGTCCGTCTTCA........................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GAGCTGGAATCGCTGAGAGTCCGTCTTCA........................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CTGGAATCGCTGAGAGTCCGTCTTCAatc........................................................................................................ | 29 | atc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CTCGTCAGGCTGTCTGAc....................................................................... | 18 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................CCCCTAGCAAACCTaatt......... | 18 | aatt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GGAGCTGGAATCGCTGAGAGTCCGTCTTCA........................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................AGCAAACCTGGAGGTGAC | 18 | 3 | 0.67 | 0.67 | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CAAGGTTCAGACCACCCCTAGCA.............. | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ...................................................................................................................................................GGCAAATCTAGTTCCA................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |