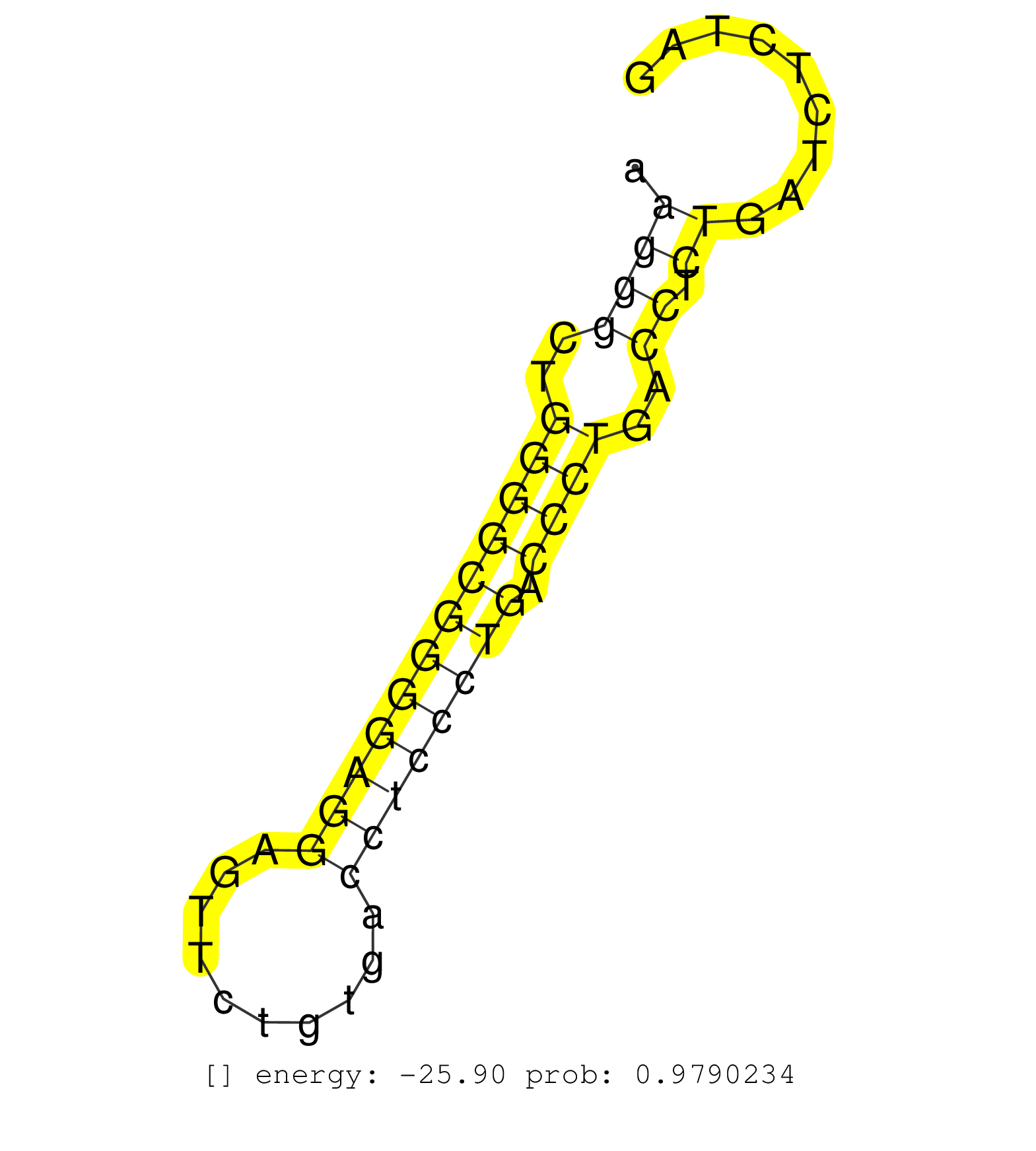
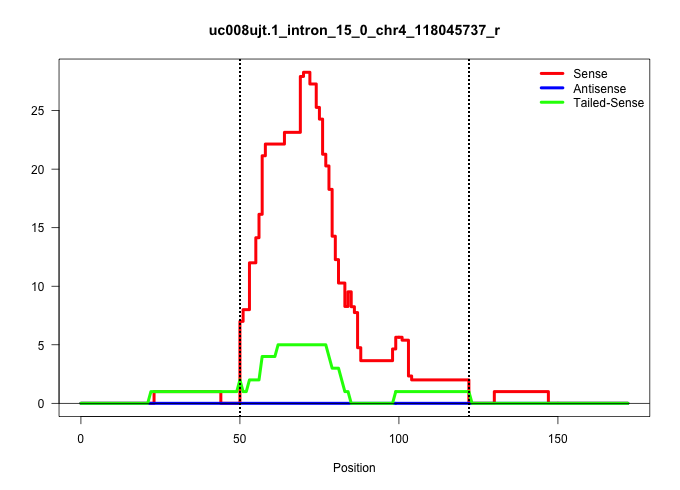
| Gene: BC059842 | ID: uc008ujt.1_intron_15_0_chr4_118045737_r | SPECIES: mm9 |
 |
 |
|
(9) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(28) TESTES |
| GAAGCGCTCCCCGGCAAAGGCTTTTGCTGCTAGAGGTTACAGATAAGAAGGTAAGTGTGGGGTCAAGGGCTGGGGCGGGGAGGAGTTCTGTGACCTCCCTGACCCTGACCTCTGATCTCTAGCTACAGCTGCTGACTTATAACTGGGCTCCAGACCTGGGTGCAGCATTGGG .................................................................((((..((((((((((((..........)))))))).))))..)).))........................................................... ................................................................65.......................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................TCAAGGGCTGGGGCGGGGAGG......................................................................................... | 21 | 1 | 15.00 | 15.00 | 12.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TCAAGGGCTGGGGCGGGGAGa......................................................................................... | 21 | a | 7.00 | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCAAGGGCTGGGGCGGGGAGG......................................................................................... | 22 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCAAGGGCTGGGGCGGGGAGtaa....................................................................................... | 24 | taa | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GTGGGGTCAAGGGCTGGGGCGGG............................................................................................. | 23 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCAAGGGCTGGGGCGGGGAG.......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGGGGCGGGGAGGAGTT..................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGGGGCGGGGAGGAGT...................................................................................... | 17 | 2 | 3.00 | 3.00 | 2.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGTGGGGTCAAGGGCTGGGGCGGGGAGG......................................................................................... | 28 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGGGCTGGGGCGGGGAGG......................................................................................... | 18 | 5 | 2.40 | 2.40 | 0.80 | 0.80 | - | - | - | - | - | - | - | - | - | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGACCCTGACCTCTGATCTCTAG.................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................GACCCTGACCTCTGATCTCTAG.................................................. | 22 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTGGG.................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCAAGGGCTGGGGCGGGGAat......................................................................................... | 22 | at | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGGGGTCAAGGGCTGGGGCGGG............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTGGGGC................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TCAAGGGCTGGGGCGGGGAG.......................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGGGGCGGGGAGGAG....................................................................................... | 16 | 11 | 1.27 | 1.27 | 1.00 | - | - | - | 0.09 | 0.18 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GCTGACTTATAACTGGG......................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGGGGTCAAGGGCTGGGGCGG.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGGGGTCAAGGGCTGGGGCGGGGA........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................TGCTGACTTATAACTGGGCTCCAGA.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGGGGCGGGGAGGAGTTC.................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGACCCTGACCTCTGATCTCT.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTG.................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGTCAAGGGCTGGGGCG............................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGTCAAGGGCTGGGGCGGa............................................................................................. | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AAGGGCTGGGGCGGGGAGG......................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTTGCTGCTAGAGGTTACAGATAAGAAGc......................................................................................................................... | 29 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGGGGTCAAGGGCTGGGGCGGGGAa.......................................................................................... | 25 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GAGTTCTGTGACCTCCCTGAC..................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGTCAAGGGCTGGGGCGGGGAGG......................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AGGAGTTCTGTGACCTCCCTGAC..................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTGGGGCGGGGAG.......................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGTCAAGGGCTGGGGCGGGG............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TCAAGGGCTGGGGCGGGGAGaat....................................................................................... | 23 | aat | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GTGGGGTCAAGGGCTGGGGC................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGGGGTCAAGGGCTGGGGCGGGGAag......................................................................................... | 26 | ag | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................TGTGGGGTCAAGGGCTGGGGCGGGG............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCAAGGGCTGGGGCGGGGAGGAa....................................................................................... | 24 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GTTCTGTGACCTCCCTGAC..................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GTGGGGTCAAGGGCTGGGGCGGGGA........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGTGGGGTCAAGGGCTGGGGCGGG............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGTGGGGTCAAGGGCTGGGGCGG.............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTGGGGCGt.............................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................GGGGTCAAGGGCTGGGGCGGG............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................TGGGGTCAAGGGCTGGGGCGGGG............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AAGGGCTGGGGCGGGGAGGAG....................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACCCTGACCTCTGATCTCTAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGACCCTGACCTCTGATCTCTAGt................................................. | 24 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................TTGCTGCTAGAGGTTACAGAT................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGTGGGGTCAAGGGCTGGGG................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................TGGGGCGGGGAGGAGTTC.................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGTGGGGTCAAGGGC...................................................................................................... | 15 | 7 | 0.43 | 0.43 | - | - | 0.14 | - | - | - | - | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGTGACCTCCCTGACC.................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GTTCTGTGACCTCCCTG....................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGTGACCTCCCTGAC..................................................................... | 15 | 17 | 0.06 | 0.06 | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAAGCGCTCCCCGGCAAAGGCTTTTGCTGCTAGAGGTTACAGATAAGAAGGTAAGTGTGGGGTCAAGGGCTGGGGCGGGGAGGAGTTCTGTGACCTCCCTGACCCTGACCTCTGATCTCTAGCTACAGCTGCTGACTTATAACTGGGCTCCAGACCTGGGTGCAGCATTGGG .................................................................((((..((((((((((((..........)))))))).))))..)).))........................................................... ................................................................65.......................................................122................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................ACAGCTGCTGACTTAgagt................................. | 19 | gagt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................CAGCTGCTGACTTAggta................................. | 18 | ggta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |