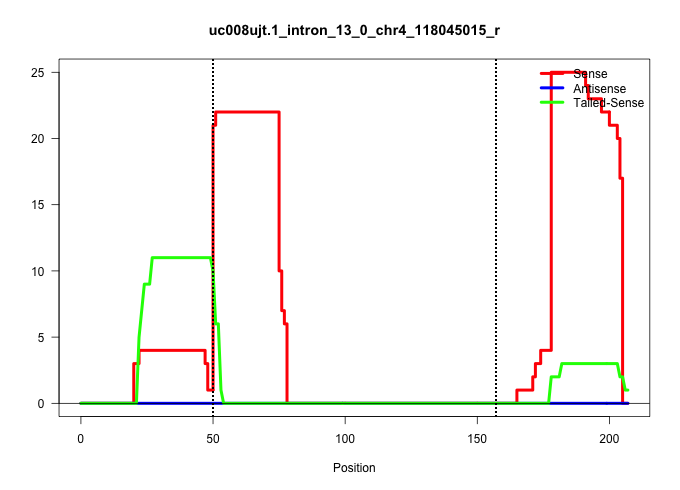
| Gene: BC059842 | ID: uc008ujt.1_intron_13_0_chr4_118045015_r | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(22) TESTES |
| GACCTACCACGGGCAGCAGTTCTTAGAAATTAAGATGACAGAACGAAAAGGTGAGGAATGTGGGCCAGGCAGCAGGGAGGGAAAATTCTGCGAAGACCTGAGTGTGACAGAGAGGAGGTCAGCTGCAAAGGCTACTACTGATGCCTCCCATCCCCAGAGCTGGAGCGCCAGATGAAGATGGAGAACCTGTTTGTAACCTGGCAGCAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TGGAGAACCTGTTTGTAACCTGGCAGC.. | 27 | 1 | 17.00 | 17.00 | - | 7.00 | 2.00 | 1.00 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGGAATGTGGGCCAGGCAGCAG.................................................................................................................................... | 25 | 1 | 12.00 | 12.00 | 7.00 | - | - | - | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGGAATGTGGGCCAGGCAGCAGGGA................................................................................................................................. | 28 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGAGAACCTGTTTGTAACCTGGCAG... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAATGTGGGCCAGGCAGCAGG................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ....................TCTTAGAAATTAAGATGACAGAACGAAA............................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTAGAAATTAAGATGACAGAACGAAAAGagc.......................................................................................................................................................... | 31 | agc | 2.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................CGCCAGATGAAGATGGAGAACCTGTT................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................ATGAAGATGGAGAACCTGTTTGTAAC.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGAGAACCTGTTTGTAACCTGGCAt... | 26 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAGAAATTAAGATGACAGAACGAAAAGt............................................................................................................................................................ | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................AATTAAGATGACAGAACGAAAAGa............................................................................................................................................................ | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGAGAACCTGTTTGTAACCTGGCA.... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTAGAAATTAAGATGACAGAACGAAAAGa............................................................................................................................................................ | 29 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................TTAGAAATTAAGATGACAGAACGAAAAt............................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTAGAAATTAAGATGACAGAACGAAAAGagt.......................................................................................................................................................... | 31 | agt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................TAGAAATTAAGATGACAGAACGAAAAGagc.......................................................................................................................................................... | 30 | agc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GAACCTGTTTGTAACCTGGCAGCAGgg | 27 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................TCTTAGAAATTAAGATGACAGAACGAA................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTAGAAATTAAGATGACAGAACGAAAAG............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGAAGATGGAGAACCTGTTTGTAACCTG....... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGAGAACCTGTTTGTAACCTGGCAGtt. | 28 | tt | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AAGATGGAGAACCTGTTT............... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AATTAAGATGACAGAACGAAAAGagct......................................................................................................................................................... | 27 | agct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGAATGTGGGCCAGGCAGCAGGG.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGAAATTAAGATGACAGAACGAAAAGa............................................................................................................................................................ | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................AGAAATTAAGATGACAGAACGAAAAGagc.......................................................................................................................................................... | 29 | agc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GACCTACCACGGGCAGCAGTTCTTAGAAATTAAGATGACAGAACGAAAAGGTGAGGAATGTGGGCCAGGCAGCAGGGAGGGAAAATTCTGCGAAGACCTGAGTGTGACAGAGAGGAGGTCAGCTGCAAAGGCTACTACTGATGCCTCCCATCCCCAGAGCTGGAGCGCCAGATGAAGATGGAGAACCTGTTTGTAACCTGGCAGCAG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|