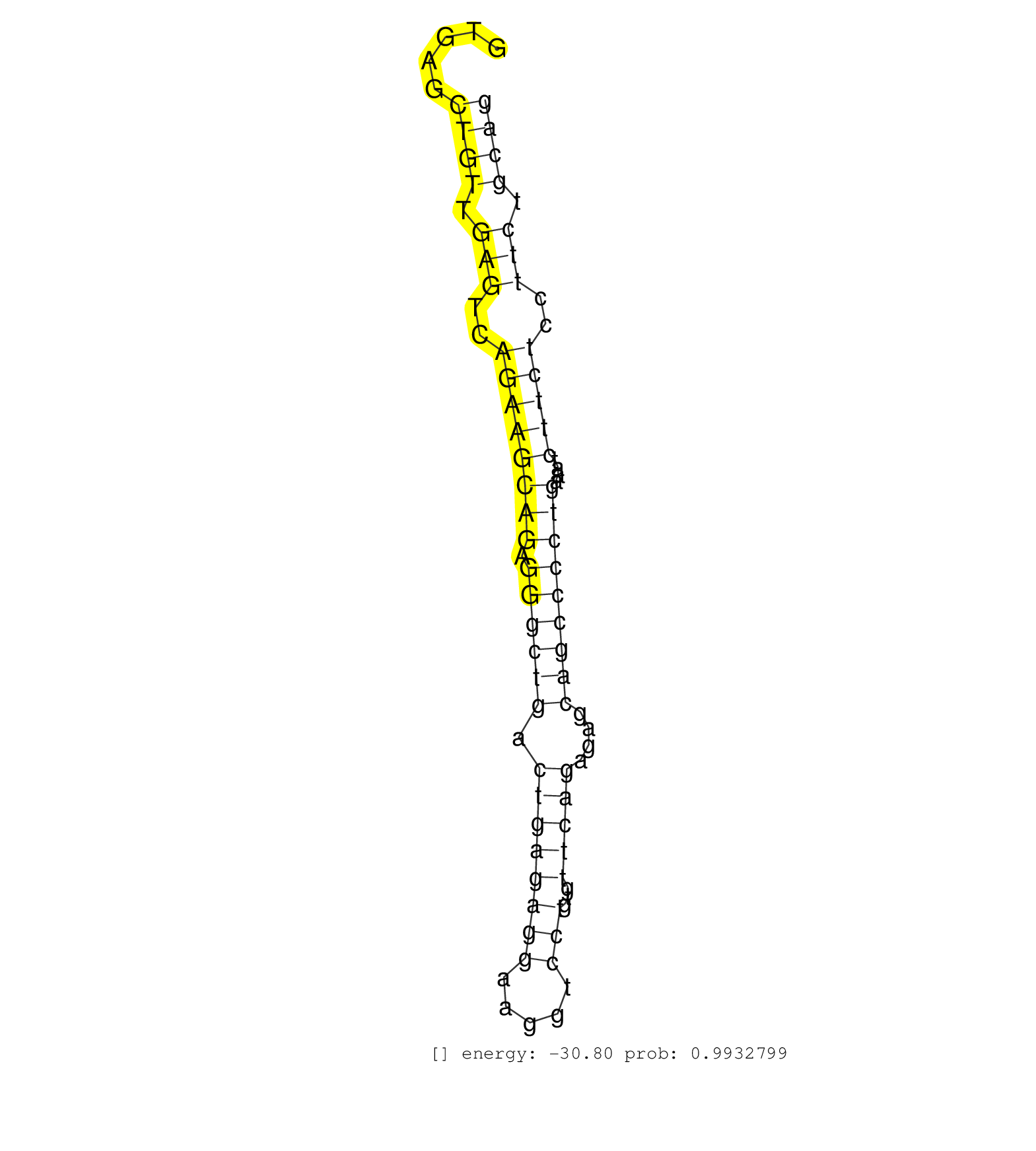
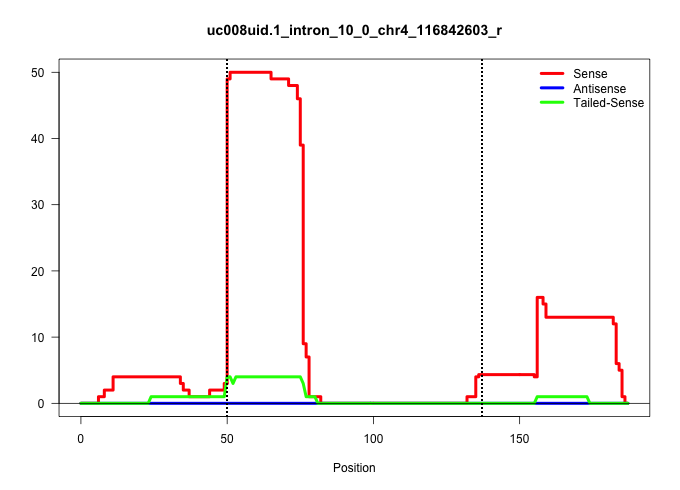
| Gene: Kif2c | ID: uc008uid.1_intron_10_0_chr4_116842603_r | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| AGAGCACAGGATCTGTGTCTGTGTCAGGAAACGCCCACTGAATAAACAAGGTGAGCTGTTGAGTCAGAAGCAGAGGGCTGACTGAGAGGAAGGTCCTGTGTTCAGAGAGCAGCCCCTGAAATCTTCTCCTTCTGCAGAACTGGCCAAGAAAGAAATCGATGTGATTTCTGTTCCCAGCAAGTGTCTC .......................................................((((.(((..((((((((.((((((.((((((((.....)))...)))))....)))))))))....)))))..))).)))).................................................. ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGG............................................................................................................... | 26 | 1 | 34.00 | 34.00 | 18.00 | 4.00 | - | 5.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGGGC............................................................................................................. | 28 | 1 | 8.00 | 8.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAG................................................................................................................ | 25 | 1 | 7.00 | 7.00 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCTGTTGAGTCAGAAGCAGAGGGC............................................................................................................. | 27 | 1 | 7.00 | 7.00 | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTTCCCAGCAAGTG.... | 27 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 1.00 | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTccag............ | 19 | ccag | 4.00 | 0.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTTCCCAGCAAGTGTC.. | 29 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGAACTGGCCAAGAAAGAAATCGA............................ | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGGG.............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGA................................................................................................................. | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGGt.............................................................................................................. | 27 | t | 2.00 | 34.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGCAGAACTGGCCAAGAAAGAAATCGA............................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGcaca............. | 18 | caca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CAGGAAACGCCCACTGAATAAACAAGaa....................................................................................................................................... | 28 | aa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................AACAAGGTGAGCTGTTGAGTCAGAAGC.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTTCCCAGCAAGT..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................TGTGATTTCTGTTCCCAGCAAGTGTC.. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGt............................................................................................................... | 26 | t | 1.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCTGTTGAGTCAGAAGCAGAGGGCTGt.......................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GGATCTGTGTCTGTGTCAGGAAACGCC........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................TGAATAAACAAGGTGAGCTGTTGAGTC.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTGTGTCTGTGTCAGGAAACGCCCA...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGTTGAGTCAGAAGCAGAGGGCTGAC......................................................................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTTCCCAGCAAGTGT... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CGATGTGATTTCTGTTCCCAGCAAGTGTCT. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTGAGCTGTTGAGTCAGAAGCAGAGG............................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCTGTTGAGTCAGAAGCAGAGG............................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGAACTGGCCAAGAAAGAAATCG............................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........TCTGTGTCTGTGTCAGGAAACGCCCAC..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......CAGGATCTGTGTCTGTGTCAGGAAACGC......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................GAACTGGCCAAGAAAGAAA................................ | 19 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ...........................................................................................................................................................TCGATGTGATTTCTG................. | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| AGAGCACAGGATCTGTGTCTGTGTCAGGAAACGCCCACTGAATAAACAAGGTGAGCTGTTGAGTCAGAAGCAGAGGGCTGACTGAGAGGAAGGTCCTGTGTTCAGAGAGCAGCCCCTGAAATCTTCTCCTTCTGCAGAACTGGCCAAGAAAGAAATCGATGTGATTTCTGTTCCCAGCAAGTGTCTC .......................................................((((.(((..((((((((.((((((.((((((((.....)))...)))))....)))))))))....)))))..))).)))).................................................. ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|