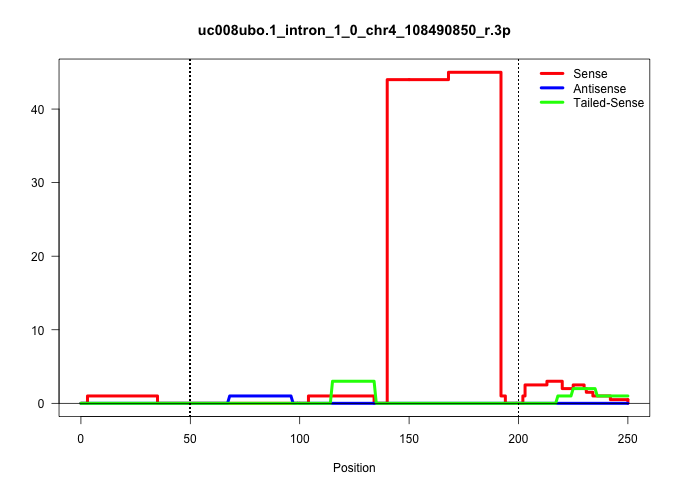
| Gene: Btf3l4 | ID: uc008ubo.1_intron_1_0_chr4_108490850_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CACCAGCTGTTCTGTATATATTTAATTCACGTTAAACTTTTATTGTTTTTAATGTTACTTAAGCAAAAAAATAGAAGTAAATTTTAGGTTGTCTGTATTCAACAGCCACATGAGATTTAGACAAAGAGAAGATAAGTATTGGAGGTGTTCTGTGAAGCTCTTTGTTCTCAGTGACTAATACTCAAATGAACTTTTTGTAGTATTGGATAGTAAAGCGCCCAAACCAGAAGACATCGATGAAGAGGATGAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................GGAGGTGTTCTGTGAAGCTCTTTGTTCTCAGTGACTAATACTCAAATGAACT.......................................................... | 52 | 1 | 44.00 | 44.00 | 44.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTTAGACAAAGAGAAGggca................................................................................................................... | 20 | ggca | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CCAAACCAGAAGACAata.............. | 18 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................CAGTGACTAATACTCAAATGAACTTT........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AGAAGACATCGATGAAGAGGATGATt | 26 | t | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - |
| ..........................................................................................................................................................................................................TTGGATAGTAAAGCGCCC.............................. | 18 | 2 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GCCACATGAGATTTAGACAAAGAGAAGATA.................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...CAGCTGTTCTGTATATATTTAATTCACGTTAA....................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGATAGTAAAGCGCCCAAACCAGAAGA................... | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGATAGTAAAGCGCCCAAACCAGAAG.................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .................................................................................................................................................................................................................................AGAAGACATCGATGAAGAGGATGAT | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGCGCCCAAACCAGAAGACATCGATGAAG........ | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...........................................................................................................................................................................................................TGGATAGTAAAGCGCCCAAACCAGAAGACAT................ | 31 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| CACCAGCTGTTCTGTATATATTTAATTCACGTTAAACTTTTATTGTTTTTAATGTTACTTAAGCAAAAAAATAGAAGTAAATTTTAGGTTGTCTGTATTCAACAGCCACATGAGATTTAGACAAAGAGAAGATAAGTATTGGAGGTGTTCTGTGAAGCTCTTTGTTCTCAGTGACTAATACTCAAATGAACTTTTTGTAGTATTGGATAGTAAAGCGCCCAAACCAGAAGACATCGATGAAGAGGATGAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................AAATAGAAGTAAATTTTAGGTTGTCTGTA......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |