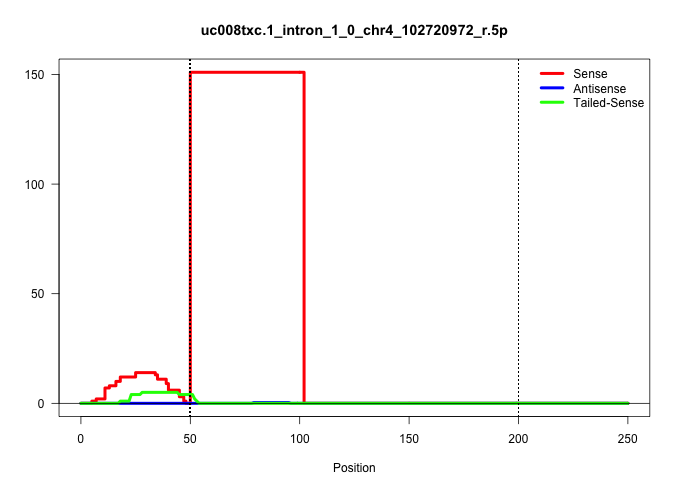
| Gene: Wdr78 | ID: uc008txc.1_intron_1_0_chr4_102720972_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| GTTCTTGTTCATACAATGAGCAATACCTAGAGACCTATAGAGGACATAAGGTTAGCTTTACTCTGATAAATGTAGAAACTGTATCTTACAGTATTTTGGGGAAACATATACAATATGTTTAATGGATATGGTAAGCTTACAACTGTGAGTTAGTAGCCCCTTGAATAAGTGTGTGTGTTGATTCCAAAGGAAAACTGTCTCTTGCTCATACTGATTTGCCCCTTTATCATCATTTTCCCAAGTACTTTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTTAGCTTTACTCTGATAAATGTAGAAACTGTATCTTACAGTATTTTGGGGA.................................................................................................................................................... | 52 | 1 | 151.00 | 151.00 | 151.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACAATGAGCAATACCTAGAGACCTATA................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................CCTAGAGACCTATAGAGGACAT........................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACAATGAGCAATACCTAGAGACCTATAG.................................................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ................TGAGCAATACCTAGAGACCTATAGAGGAC............................................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................TACCTAGAGACCTATAGAGGACATAAGtt...................................................................................................................................................................................................... | 29 | tt | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .......TTCATACAATGAGCAATACCTAGAGACC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACAATGAGCAATACCTAGAGACC....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................AGCAATACCTAGAGACCTATAGAGGACATA.......................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................AGAGACCTATAGAGGACATAAGtttg.................................................................................................................................................................................................... | 26 | tttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................AGCAATACCTAGAGACCTATAGAtgac............................................................................................................................................................................................................. | 27 | tgac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................TACCTAGAGACCTATAGAGGACATAAGttt..................................................................................................................................................................................................... | 30 | ttt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............CAATGAGCAATACCTAGAGACCTATAG.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................AGCAATACCTAGAGACCTATAGAGGAC............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTTCATACAATGAGCAATACCTAGAGAC........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GTTCTTGTTCATACAATGAGCAATACCTAGAGACCTATAGAGGACATAAGGTTAGCTTTACTCTGATAAATGTAGAAACTGTATCTTACAGTATTTTGGGGAAACATATACAATATGTTTAATGGATATGGTAAGCTTACAACTGTGAGTTAGTAGCCCCTTGAATAAGTGTGTGTGTTGATTCCAAAGGAAAACTGTCTCTTGCTCATACTGATTTGCCCCTTTATCATCATTTTCCCAAGTACTTTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................TGTATCTTACAGTATTT.......................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |