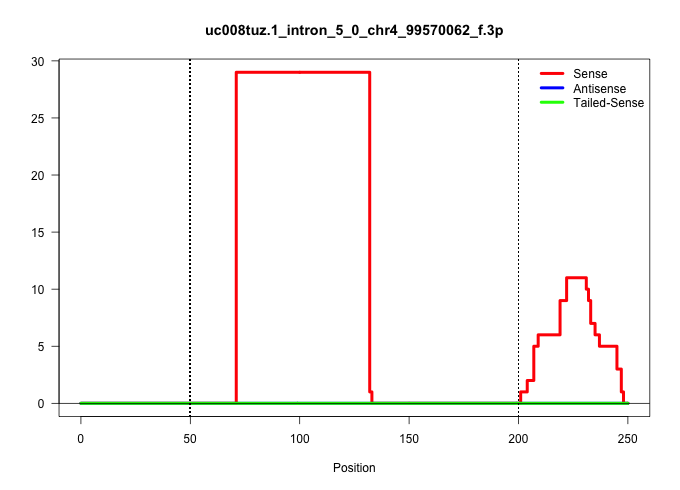
| Gene: BC020077 | ID: uc008tuz.1_intron_5_0_chr4_99570062_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(13) TESTES |
| CCACAGTCCATGGCCTTGCTCTAGGGCTACGTACAAGATAGAAAAGAAAAGAATCTAAAAACAGTGAAAGATTAATAAACTATAACTCAATTGATAAATGTTGACTTAAATCAGATGATATCTAAAGTGCCCCCCTCAGAAGAAAGATTTAAATCATTAAGATTTTTGGTGTTTATATTCTTTAATTCTTTTGCTAAAAGGTGTTTGGATGGACTGGTGAACTAGAAGCAGGAATTTATTGGTTAATTCC .......................................................................................................((((..(((....))).....))))........(((((((((((..((((.(((((((...)))))))))))...))))....)))))))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................TTAATAAACTATAACTCAATTGATAAATGTTGACTTAAATCAGATGATATCT............................................................................................................................... | 52 | 1 | 29.00 | 29.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AACTAGAAGCAGGAATTTATTGGTTA..... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................GATGGACTGGTGAACTAGAAGCAGG.................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................GATGGACTGGTGAACTAGAAGCAGGA................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TAGAAGCAGGAATTTATTGGTTAATT.. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGTTTGGATGGACTGGTGAACTAGAAGCAGG.................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TTGGATGGACTGGTGAACTAGAAGCAG................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................GATGGACTGGTGAACTAGAAGCAGGAAT............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................................TAGAAGCAGGAATTTATTGGTTAAT... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................TGGACTGGTGAACTAGAAGCAGGAATTT............. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AACTAGAAGCAGGAATTTATTGGTTAAT... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| CCACAGTCCATGGCCTTGCTCTAGGGCTACGTACAAGATAGAAAAGAAAAGAATCTAAAAACAGTGAAAGATTAATAAACTATAACTCAATTGATAAATGTTGACTTAAATCAGATGATATCTAAAGTGCCCCCCTCAGAAGAAAGATTTAAATCATTAAGATTTTTGGTGTTTATATTCTTTAATTCTTTTGCTAAAAGGTGTTTGGATGGACTGGTGAACTAGAAGCAGGAATTTATTGGTTAATTCC .......................................................................................................((((..(((....))).....))))........(((((((((((..((((.(((((((...)))))))))))...))))....)))))))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................AATAAACTATAACtgat.................................................................................................................................................................... | 17 | tgat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |