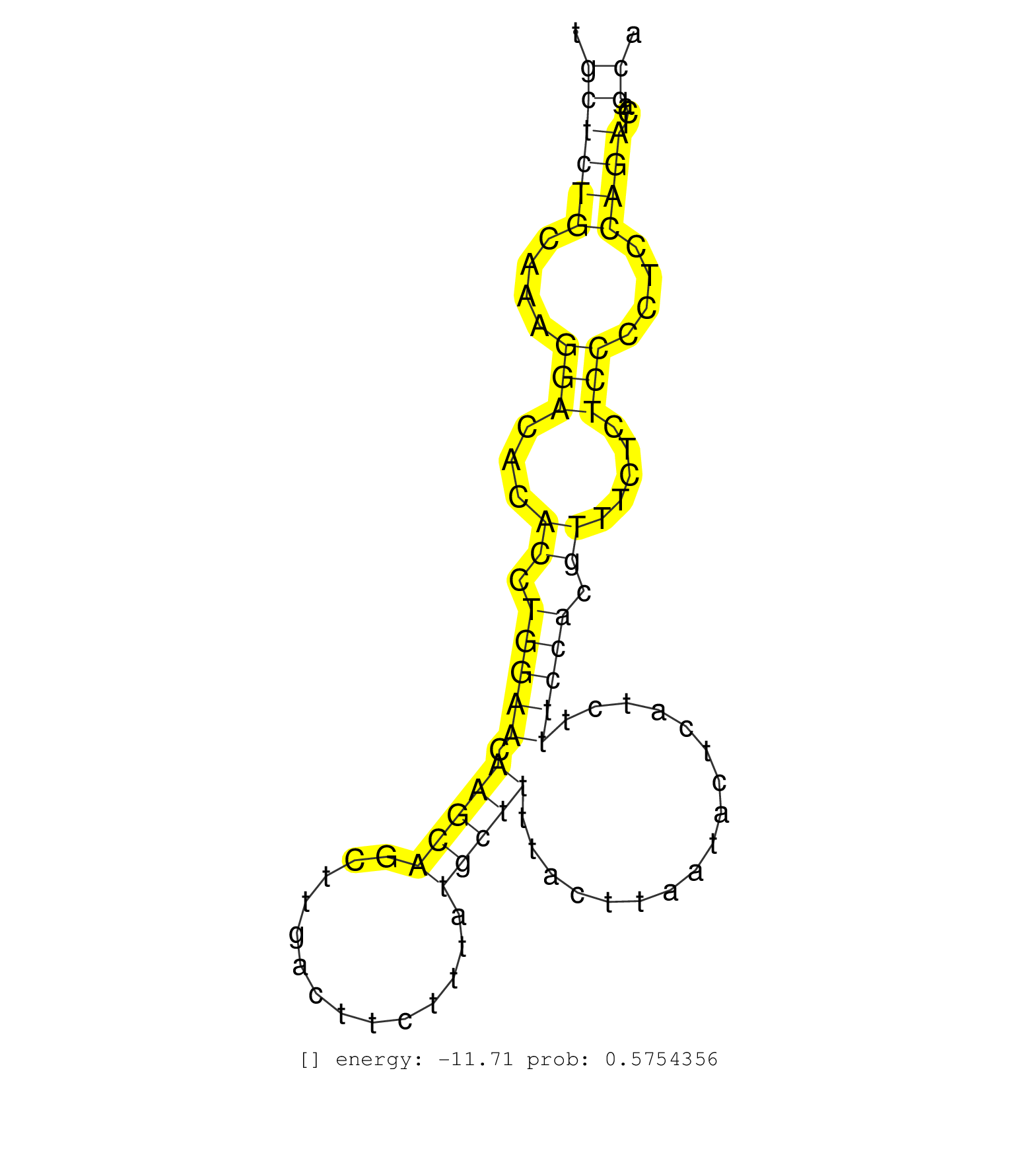
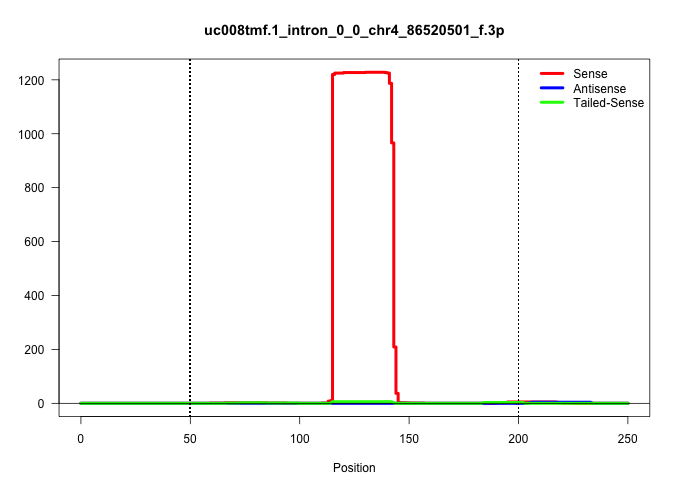
| Gene: Asah3l | ID: uc008tmf.1_intron_0_0_chr4_86520501_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(29) TESTES |
| TGTTATCCGTGTCGTGCCACCCAACCCCCCACACCCGCCCTGCGCTTGCTCTGTGCGGCTGTAGTTTAGAAAGTGCAGAGCTTGTACAAGATGCATGGTCACGTTCACCCTGCTCTGCAAAGGACACACCTGGAACAAGCAGCTTGACTTCTTTATGCTTTTACTTAATACTCATCTTTCCACGTTTCTCTCCCCTCCAGATCAGCAACGTCTTGTTTTTCATTTTACCTCCCATCTGCATGTGCTTGTT ...............................................................................................................((((((....(((...((.(((((.(((((..............))))).................))))).)).....)))....))))...))............................................ ..............................................................................................................111.............................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGC........................................................................................................... | 28 | 1 | 842.00 | 842.00 | 224.00 | 229.00 | 143.00 | 124.00 | 41.00 | 28.00 | 17.00 | 7.00 | 6.00 | 4.00 | - | 3.00 | 2.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAG............................................................................................................ | 27 | 1 | 260.00 | 260.00 | 58.00 | 65.00 | 45.00 | 37.00 | 16.00 | 11.00 | 6.00 | 5.00 | 1.00 | 2.00 | 5.00 | 2.00 | 1.00 | - | 1.00 | 2.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGCT.......................................................................................................... | 29 | 1 | 171.00 | 171.00 | 65.00 | 51.00 | 24.00 | 29.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGCTT......................................................................................................... | 30 | 1 | 34.00 | 34.00 | 15.00 | 11.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCA............................................................................................................. | 26 | 1 | 31.00 | 31.00 | 10.00 | 3.00 | 10.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................TCTGCAAAGGACACACCTGGAACAAGCA............................................................................................................. | 28 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GCAAAGGACACACCTGGAACAAGCAGC........................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TCCAGATCAGCAACGTCTTGTTTTT.............................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGt........................................................................................................... | 28 | t | 2.00 | 260.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGC.............................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTCTCTCCCCTCCAGtac............................................... | 19 | tac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTAGTTTAGAAAGTGCAGAGCTTGT..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGGAACAAGCAGCTTGACTTCTTTATG............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAGAAAGTGCAGAGCTTGTACAAGATGC............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCCCCTCCAGATCAGCAACGTCTTGTT................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................TCCAGATCAGCAACGTCTTGTTTTTCATT.......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................AGAAAGTGCAGAGCcaca..................................................................................................................................................................... | 18 | caca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCTGCAAAGGACACACCTGGAACAAGCAG............................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CTGCAAAGGACACACCTGGAACAAGCAG............................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTCTCTCCCCTCCAGgat............................................... | 19 | gat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGCc.......................................................................................................... | 29 | c | 1.00 | 842.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CTGCAAAGGACACACCTGGAACAAGCA............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GGACACACCTGGAACAAGCAGCTTGA....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAGa........................................................................................................... | 28 | a | 1.00 | 260.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAcc........................................................................................................... | 28 | cc | 1.00 | 31.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GTACAAGATGCATGGTCAC.................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................AGGACACACCTGGAACAAGCAGCTTGACT..................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGCAGAGCTTGTACAAGATGCATGGTa...................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTCTCTCCCCTCCAGtct............................................... | 19 | tct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCCCCTCCAGATCAGCAACGTCTTGTTT................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................TGCTCTGCAAAGGACACACCTGGAACAAG............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GCAAAGGACACACCTGGAACAAGCAGCT.......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGGACACACCTGGAACAAGCAGCTTG........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGCAAAGGACACACCTGGAACAAGCAtct.......................................................................................................... | 29 | tct | 1.00 | 31.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTTATCCGTGTCGTGCCACCCAACCCCCCACACCCGCCCTGCGCTTGCTCTGTGCGGCTGTAGTTTAGAAAGTGCAGAGCTTGTACAAGATGCATGGTCACGTTCACCCTGCTCTGCAAAGGACACACCTGGAACAAGCAGCTTGACTTCTTTATGCTTTTACTTAATACTCATCTTTCCACGTTTCTCTCCCCTCCAGATCAGCAACGTCTTGTTTTTCATTTTACCTCCCATCTGCATGTGCTTGTT ...............................................................................................................((((((....(((...((.(((((.(((((..............))))).................))))).)).....)))....))))...))............................................ ..............................................................................................................111.............................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................AACGTCTTGTTTTTCATTTTACCTCCCA................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAACGTCTTGTTTTTCATTTTACCTCCCA................ | 29 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |