
| Gene: Akna | ID: uc008tgb.1_intron_1_0_chr4_63031687_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(6) TESTES |
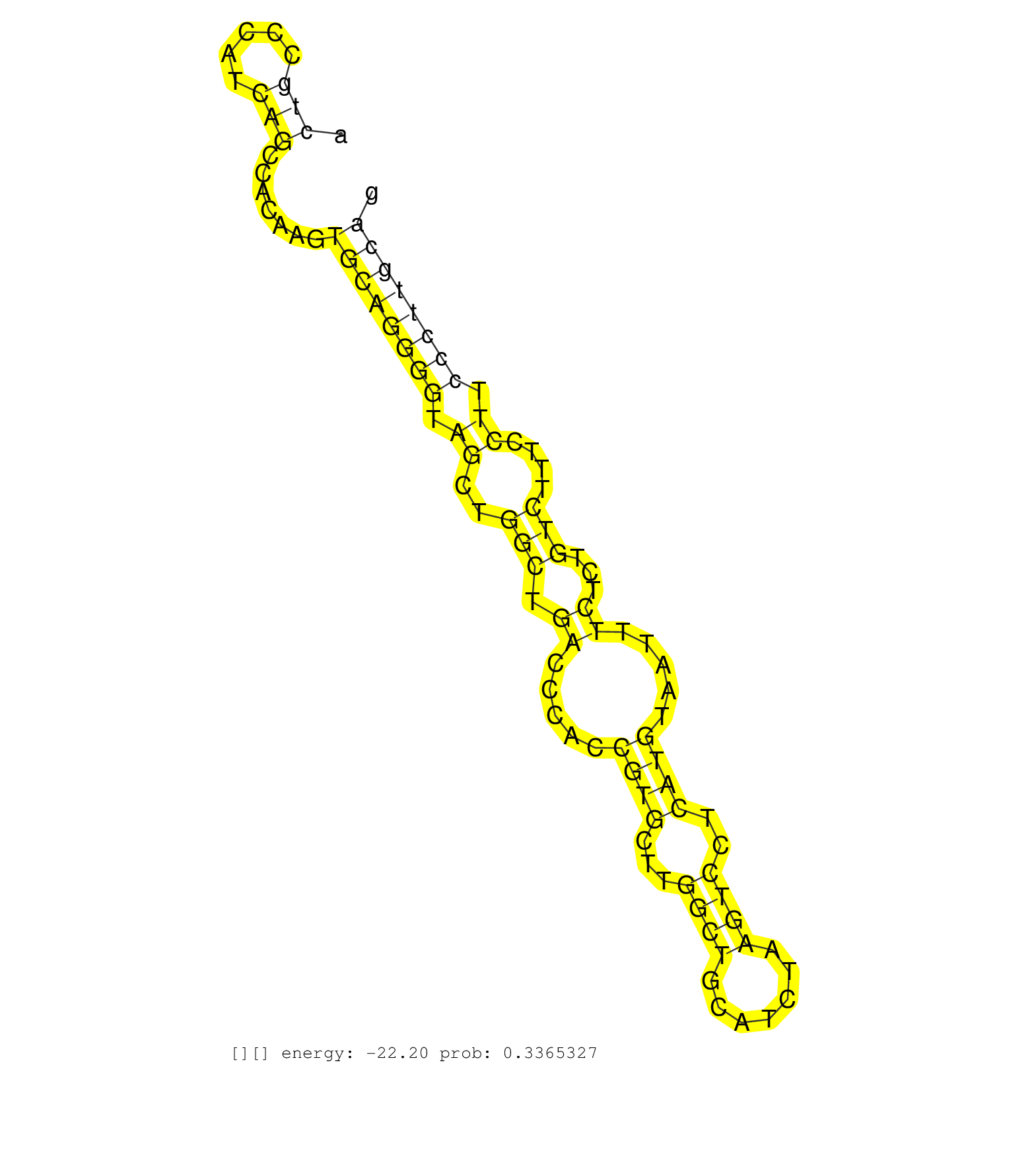
| TGAAGTCAAGTCCTCTGCAGAAGCAGATGGCTCCAGCTCAGGTCCCTCTGGTAGGAATGAGCCCTCAGCATCTTTCTTCAGCCACGGGGCTTGGCCAGACCTTGGGGATGCTGTAAGCTCCGCTAGAGCAAAATAAATGCAACAGAGGCTCAACATCCCAGAGGGATGCCCTCCCTAGGGTGGTCCTCCACTGCCCATCAGCCACAAGTGCAGGGGTAGCTGGCTGACCCACCGTGCTTGGCTGCATCTAAGTCCTCATGTAATTTCTCTGTCTTTCCTTCCCTTGCAGAGAAGAACACCCCCAAAAAGCCTTCCACCCCCATCCTCAAACGGAAGAAC ..............................................................................................................................................................................................(((.....))).......((((((((.((..(((.((.....((((...((((.......))))..)))).....))...)))....)).))))))))................................................... .............................................................................................................................................................................................190................................................................................................289................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................CCCATCAGCCACAAGTGCAGGGGTAGCTGGCTGACCCACCGTGCTTGGCTGC.............................................................................................. | 52 | 1 | 73.00 | 73.00 | 73.00 | - | - | - | - | - |
| ..................................................GTAGGAATGAGCCCTCAGCATCTTTC....................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| ..................................................GTAGGAATGAGCCCTCAGCATCTTTa....................................................................................................................................................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - |
| .......AAGTCCTCTGCAGAAGCAGATGG..................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ..................................................GTAGGAATGAGCCCTCAGCATCTTTCTT..................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ........................................................................................................GGGATGCTGTAAGCT............................................................................................................................................................................................................................ | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | 0.12 |
| TGAAGTCAAGTCCTCTGCAGAAGCAGATGGCTCCAGCTCAGGTCCCTCTGGTAGGAATGAGCCCTCAGCATCTTTCTTCAGCCACGGGGCTTGGCCAGACCTTGGGGATGCTGTAAGCTCCGCTAGAGCAAAATAAATGCAACAGAGGCTCAACATCCCAGAGGGATGCCCTCCCTAGGGTGGTCCTCCACTGCCCATCAGCCACAAGTGCAGGGGTAGCTGGCTGACCCACCGTGCTTGGCTGCATCTAAGTCCTCATGTAATTTCTCTGTCTTTCCTTCCCTTGCAGAGAAGAACACCCCCAAAAAGCCTTCCACCCCCATCCTCAAACGGAAGAAC ..............................................................................................................................................................................................(((.....))).......((((((((.((..(((.((.....((((...((((.......))))..)))).....))...)))....)).))))))))................................................... .............................................................................................................................................................................................190................................................................................................289................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................GCAACAGAGGCTCAAccca.......................................................................................................................................................................................... | 19 | ccca | 1.00 | 0.00 | - | - | - | 1.00 | - | - |