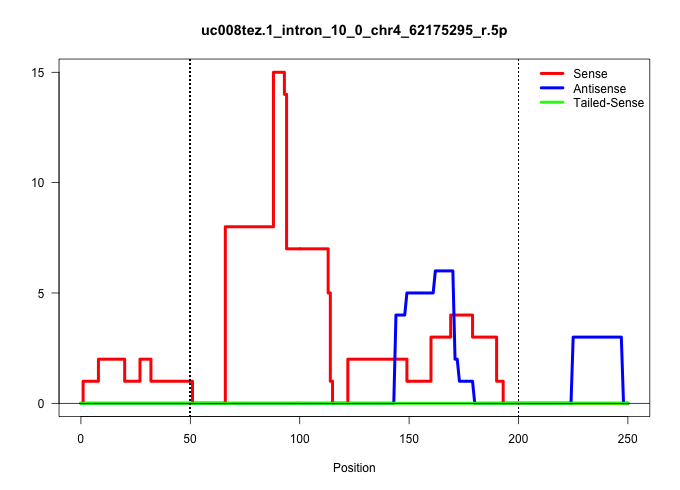
| Gene: Alad | ID: uc008tez.1_intron_10_0_chr4_62175295_r.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| CCGGCCTCTGGGAAGTGGCGGGTTGAAGGGGGACGGGACCCCAGAACAAGGTGAGGTGCACCCCTATGGCGCTGTGTTCCCGAAGCTCTGACGCTGTAGCGGAGACGCTGTCCCTATGCACCTACTAGAGTGGGCGGGATTCTTGCCGCTACAAGGTCTTGCTGCGTCCGGGATTCGGGACTCAAAGATGGGCTCCACCCGCCCCCTTTCTCGGAGGCCTCAGGGGTAGCCTCGGTTTCTGCAGAGCGCG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................TGGCGCTGTGTTCCCGAAGCTCTGACGC............................................................................................................................................................ | 28 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGACGCTGTAGCGGAGACGCTGTCCC........................................................................................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TACTAGAGTGGGCGGGATTCTTGCCGC..................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GCTGCGTCCGGGATTCGGGACTCAAAGATG............................................................ | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGACGCTGTAGCGGAGACGCTGTCC......................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .CGGCCTCTGGGAAGTGGCG...................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................TGACGCTGTAGCGGAGACGCTGTCCCT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........TGGGAAGTGGCGGGTTGAAGGGGG.......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................TGGCGCTGTGTTCCCGAAGCTCTGACG............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................TACAAGGTCTTGCTGCGTCCGGGATTCGGG....................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GGGATTCGGGACTCAAAGATGGGC......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................GGGGGACGGGACCCCAGAACAAGG....................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CCGGCCTCTGGGAAGTGGCGGGTTGAAGGGGGACGGGACCCCAGAACAAGGTGAGGTGCACCCCTATGGCGCTGTGTTCCCGAAGCTCTGACGCTGTAGCGGAGACGCTGTCCCTATGCACCTACTAGAGTGGGCGGGATTCTTGCCGCTACAAGGTCTTGCTGCGTCCGGGATTCGGGACTCAAAGATGGGCTCCACCCGCCCCCTTTCTCGGAGGCCTCAGGGGTAGCCTCGGTTTCTGCAGAGCGCG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................GCCGCTACAAGGTCTTGCTGCGTCCGG............................................................................... | 27 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GTAGCCTCGGTTTCTGCAGAGCG.. | 23 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CGCTACAAGGTCcaac............................................................................................ | 16 | caac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................TACAAGGTCTTGCTGCGTCCGGGA............................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................TGCGTCCGGGATTCGGGA...................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |