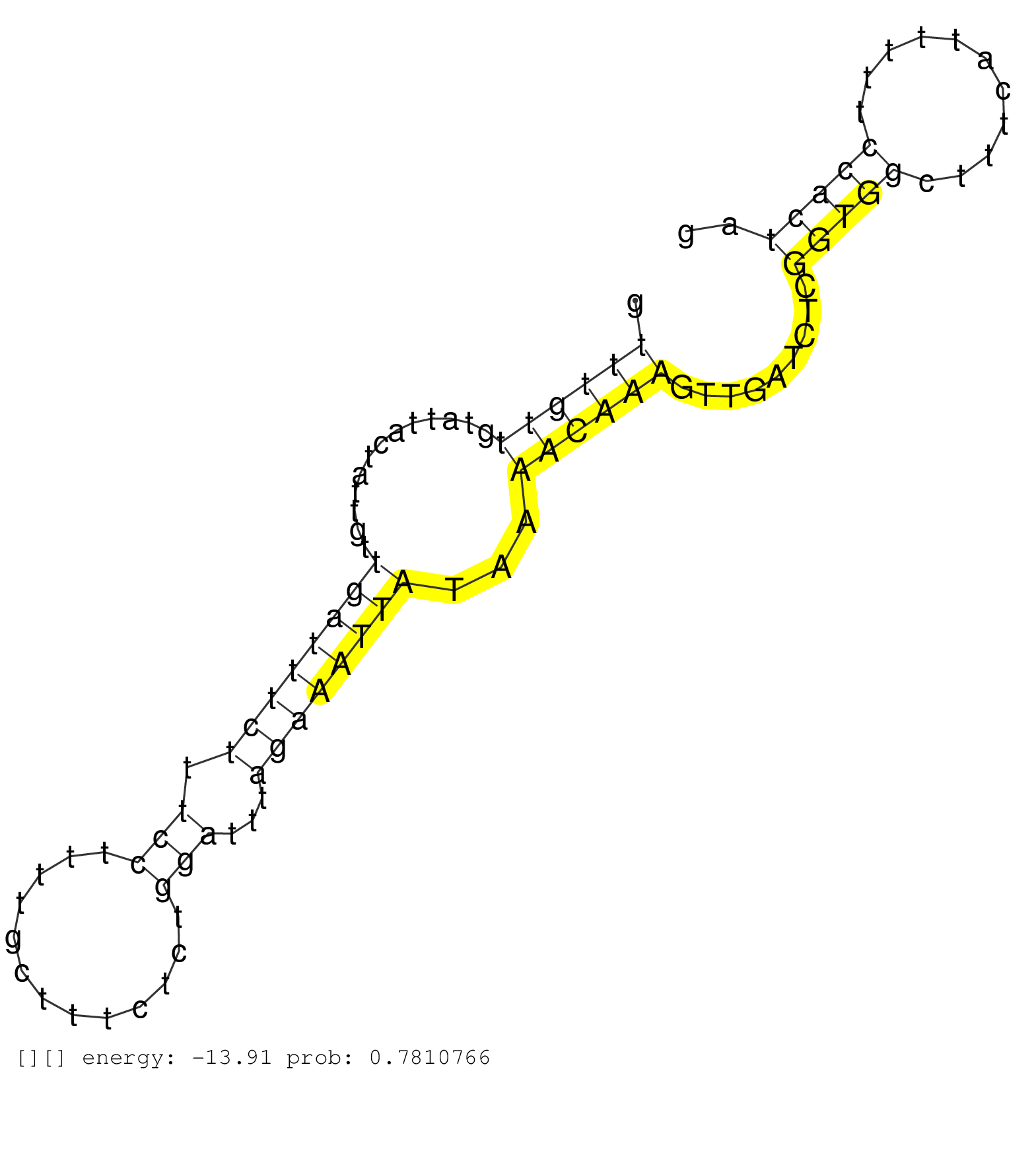
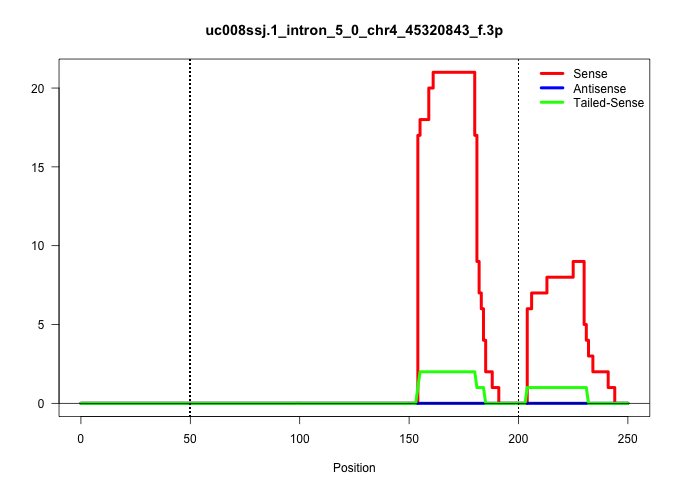
| Gene: Rg9mtd3 | ID: uc008ssj.1_intron_5_0_chr4_45320843_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(18) TESTES |
| TCTCGTACGTCTGGCTTTGTTTCCCGTCTAACTCAGAGCTGCTTTGGACAGCACCTGCGTTTTTTCAGATCTGCCAGCAGGCTTTAAGTTGTGTTGTTGTGTTTGTTGTATTACTATTGTTGATTTCTTTCCTTTTGCTTTCTCTGGATTTAGAAATTATAAAACAAAGTTGATCTCGGTGGCTTTCATTTTTCCACTAGCCCTTGAAGACATTGATCAGAGCACAGTTTACGTTATTGGTGGACTTGTA .....................................................................................................((((((.............((((((((.(((.............)))...))))))))...)))))).........(((((...........))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGTG..................................................................... | 27 | 1 | 8.00 | 8.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 |
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGT...................................................................... | 26 | 1 | 5.00 | 5.00 | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGACATTGATCAGAGCACAGTTT.................... | 26 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGTGGCTT................................................................. | 31 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGTGGCT.................................................................. | 30 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................TAAAACAAAGTTGATCTCGGTGGCTTTCA.............................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TAAAACAAAGTTGATCTCGGTGG.................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AGTTTACGTTATTGGTGGA...... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGACATTGATCAGAGCACAGTTTA................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGTt..................................................................... | 27 | t | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................AAGACATTGATCAGAGCACAGTTTACGT................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................TGATCAGAGCACAGTTTACGTTATTGGT......... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................ATTATAAAACAAAGTTGATCTCGGTGcctt................................................................. | 30 | cctt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGACATTGATCAGAGCACAGTTTAC.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................ATTATAAAACAAAGTTGATCTCGGTGG.................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGAAGACATTGATCAGAGCACAGTTTtt.................. | 28 | tt | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAACAAAGTTGATCTCGGTGGCTTTCATTT........................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................AATTATAAAACAAAGTTGATCTCGGTGGC................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTCGTACGTCTGGCTTTGTTTCCCGTCTAACTCAGAGCTGCTTTGGACAGCACCTGCGTTTTTTCAGATCTGCCAGCAGGCTTTAAGTTGTGTTGTTGTGTTTGTTGTATTACTATTGTTGATTTCTTTCCTTTTGCTTTCTCTGGATTTAGAAATTATAAAACAAAGTTGATCTCGGTGGCTTTCATTTTTCCACTAGCCCTTGAAGACATTGATCAGAGCACAGTTTACGTTATTGGTGGACTTGTA .....................................................................................................((((((.............((((((((.(((.............)))...))))))))...)))))).........(((((...........))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|