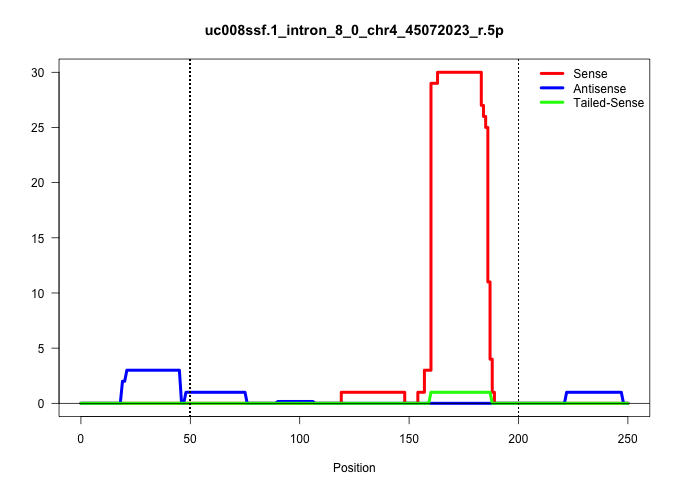
| Gene: Fbxo10 | ID: uc008ssf.1_intron_8_0_chr4_45072023_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(11) TESTES |
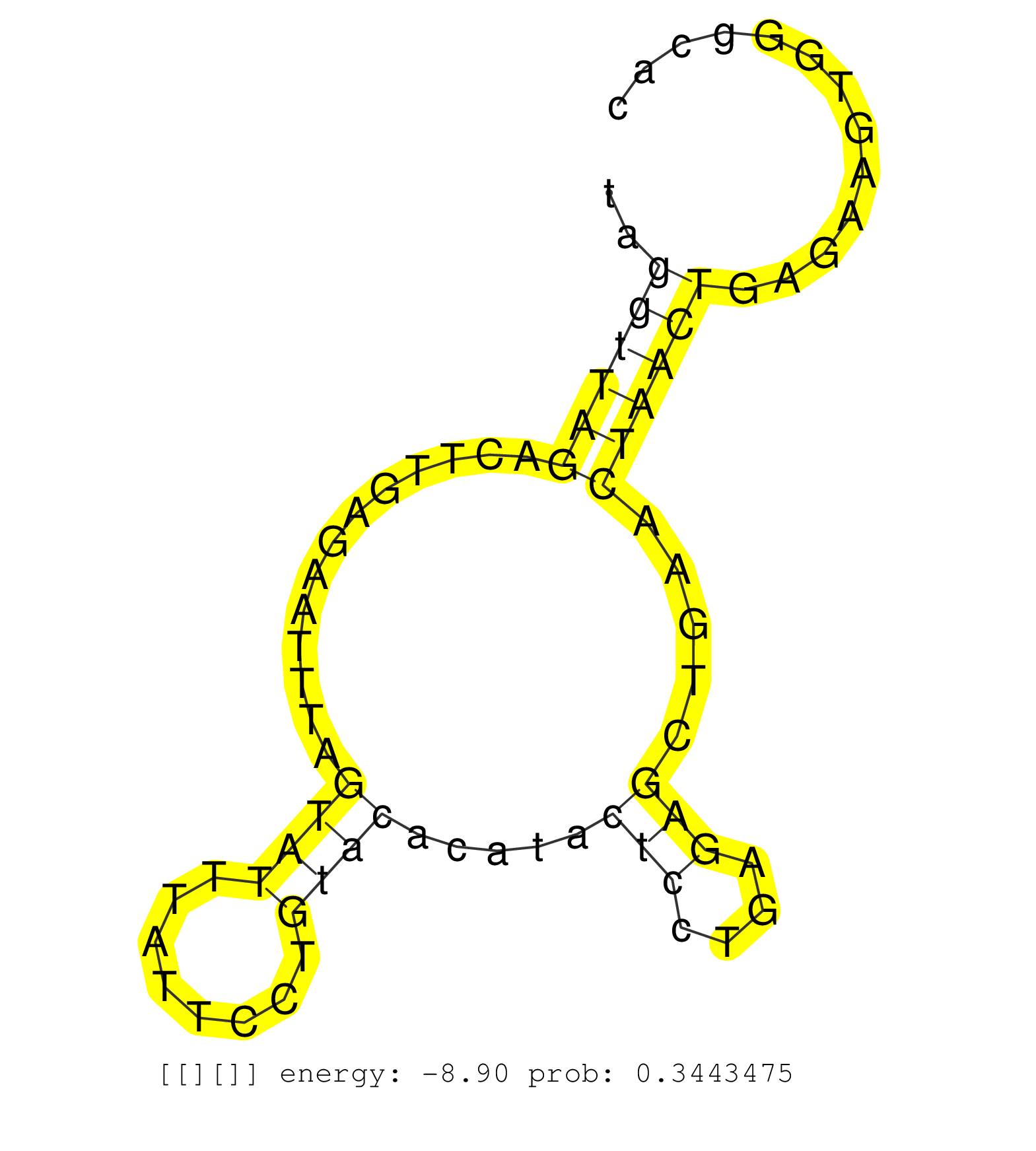
| TGTGCAATCTTGTCTTCATGCCAGCCTGGTTCTCACCAATCATGTATAAGGTGGGTGCACACCTGATACCATCCAATCCTTACCCCACGGCCTTTCTCTCAGTGCCTTTCCCTTTAGGTTAGACTTGAGAATTTAGTATTTATTCCTGTACACATACTCCTGAGAGCTGAACTAACTGAGAAGTGGGCACACAGGTCAGTATGGTCCAGAGTTATGCTTGGTGCCTTAATTCCTTCAAAACTGCCTTAGA ....................................................................................................................((((((.............((((........)))).....(((....))).....))))))......................................................................... ..................................................................................................................115........................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAGTGG................................................................ | 26 | 1 | 14.00 | 14.00 | 2.00 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAGTGGG............................................................... | 27 | 1 | 7.00 | 7.00 | - | 2.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAGTGGGC.............................................................. | 28 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TCCTGAGAGCTGAACTAACTGAGAAGT.................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAGTG................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAGTGGac.............................................................. | 28 | ac | 1.00 | 14.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TACTCCTGAGAGCTGAACTAACTGAGAAG................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................TAGACTTGAGAATTTAGTATTTATTCCTG...................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TCCTGAGAGCTGAACTAACTGAGAAG................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GAGCTGAACTAACTGAGAAGTGGGCA............................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGAGAGCTGAACTAACTGAGAAG................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| TGTGCAATCTTGTCTTCATGCCAGCCTGGTTCTCACCAATCATGTATAAGGTGGGTGCACACCTGATACCATCCAATCCTTACCCCACGGCCTTTCTCTCAGTGCCTTTCCCTTTAGGTTAGACTTGAGAATTTAGTATTTATTCCTGTACACATACTCCTGAGAGCTGAACTAACTGAGAAGTGGGCACACAGGTCAGTATGGTCCAGAGTTATGCTTGGTGCCTTAATTCCTTCAAAACTGCCTTAGA ....................................................................................................................((((((.............((((........)))).....(((....))).....))))))......................................................................... ..................................................................................................................115........................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................GCCAGCCTGGTTCTCACCAATCATGTA............................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................GCCTTAATTCCTTCAAAACTGCCTTA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCAGTATGGTCCAGAGTTATGCTTGGTa............................ | 28 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................CAGCCTGGTTCTCACCAATCATGTA............................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................AGGTGGGTGCACACCTGATACCATCCAA.............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................TCTCTCAGTGCCTTTCCCTTTAGGTTAtt................................................................................................................................. | 29 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................CCAATCCTTACCCCctta.................................................................................................................................................................... | 18 | ctta | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................CCTTTCTCTCAGTGCCT............................................................................................................................................... | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 |