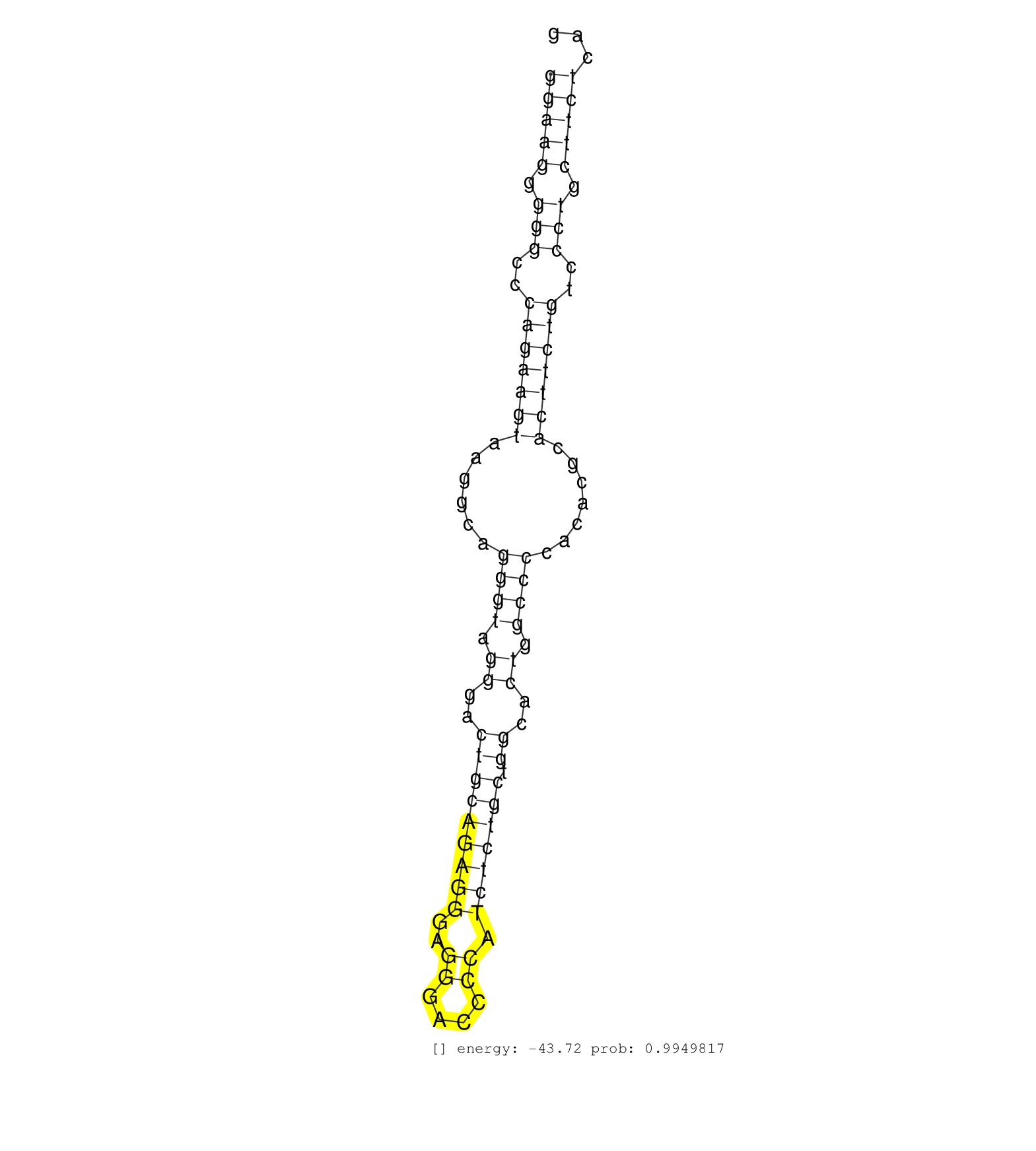

| Gene: Tpm2 | ID: uc008sqb.1_intron_5_0_chr4_43532379_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(1) PIWI.mut |
(11) TESTES |
| GCCTTGCAAAAGCTGGAGGAGGCTGAGAAAGCCGCGGATGAGAGCGAGAGGTGAGGAAGGGGGCCCAGAAGTAAGGCAGGGTAGGGACTGCAGAGGGAGGGACCCCATCTCTGCTGGCACTGGCCCCACACGCACTTCTGTCCCTGCTTCTCAGAGGAATGAAGGTCATTGAAAACCGGGCCATGAAGGATGAGGAAAAGATGG ......................................................(((((.(((..(((((((......((((.((..(((((((((..((....)).))))))).))..)).)))).......)))))))..))).)))))..................................................... ......................................................55.................................................................................................154................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................AAACCGGGCCATGAAGGATGAG.......... | 22 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GAAGGATGAGGAAAAGATGG | 20 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GCCGCGGATGAGAGCGAGAGagga...................................................................................................................................................... | 24 | agga | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| .............................AGCCGCGGATGAGAGCGAGAG.......................................................................................................................................................... | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAGGTCATTGAAAACCGGGCCATGAAGG............... | 28 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................AAAGCCGCGGATGAGAGCGAGAGagg....................................................................................................................................................... | 26 | agg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....TGCAAAAGCTGGAGGAGGCTGAGAA............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................TTCTGTCCCTGCTTCTCAGAa................................................ | 21 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGAAAACCGGGCCATGAAGGATGAGG......... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GTCATTGAAAACCGGGCCATGAA................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGAAGGATGAGGAAAAGATG. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................GAGAAAGCCGCGGATGAGAGCGAGAGagg....................................................................................................................................................... | 29 | agg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................AGAGGGAGGGACCgcag................................................................................................ | 17 | gcag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................ATTGAAAACCGGGCCATGAAGGATG............ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................GCGGATGAGAGCGAGAGagga...................................................................................................................................................... | 21 | agga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAAGGGGGCCCAGAAGTAA.................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| GCCTTGCAAAAGCTGGAGGAGGCTGAGAAAGCCGCGGATGAGAGCGAGAGGTGAGGAAGGGGGCCCAGAAGTAAGGCAGGGTAGGGACTGCAGAGGGAGGGACCCCATCTCTGCTGGCACTGGCCCCACACGCACTTCTGTCCCTGCTTCTCAGAGGAATGAAGGTCATTGAAAACCGGGCCATGAAGGATGAGGAAAAGATGG ......................................................(((((.(((..(((((((......((((.((..(((((((((..((....)).))))))).))..)).)))).......)))))))..))).)))))..................................................... ......................................................55.................................................................................................154................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................CCTGCTTCTCAGAGGAATGAAGGTCA.................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................AGAGCGAGAGGTGAgttt...................................................................................................................................................... | 18 | gttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................TCCCTGCTTCTCAGAGGAATGAAGGTCA.................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................GGGTAGGGACTGCAaata................................................................................................................ | 18 | aata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |