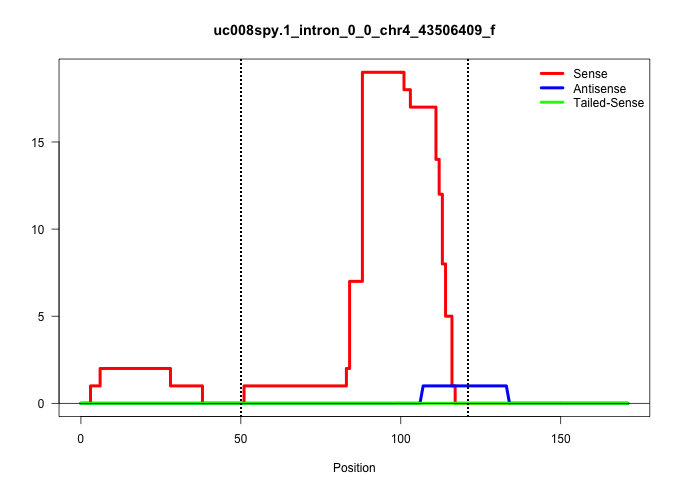
| Gene: LOC622404 | ID: uc008spy.1_intron_0_0_chr4_43506409_f | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(12) TESTES |
| TTTTGGAGTCCTTGGGGAGCGCCCCAGCGCCGATCTGGGGGCACACCCAGGTACCTGCGAGAGCTGCTGGAGGGAGGCTGGGACTGCCTAGTAGAAACTGACCGATGGCTTTAATCCACAGAACGGGGCTCCCAGGTCAGTCCGGGAACCACCGAACCTCGGCGGCAGCCG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TAGTAGAAACTGACCGATGGCTTTA.......................................................... | 25 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAGTAGAAACTGACCGATGGCTTTAATC....................................................... | 28 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................TAGTAGAAACTGACCGATGGCTTTAA......................................................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTAGTAGAAACTGACCGATGGCTT............................................................ | 27 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTAGTAGAAACTGACCGATGGCTTT........................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................TACCTGCGAGAGCTGCTGGAGGGAG............................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................CTGCCTAGTAGAAACTGA...................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...TGGAGTCCTTGGGGAGCGCCCCAGC............................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................GCTGGGACTGCCTAGTAGAAACTGACC.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......AGTCCTTGGGGAGCGCCCCAGCGCCGATCTGG..................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................TAGTAGAAACTGACCGATGGCTTTAATCC...................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TTTTGGAGTCCTTGGGGAGCGCCCCAGCGCCGATCTGGGGGCACACCCAGGTACCTGCGAGAGCTGCTGGAGGGAGGCTGGGACTGCCTAGTAGAAACTGACCGATGGCTTTAATCCACAGAACGGGGCTCCCAGGTCAGTCCGGGAACCACCGAACCTCGGCGGCAGCCG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................GAGAGCTGCTGGAGGcga.................................................................................................. | 18 | cga | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ...........................................................................................................GCTTTAATCCACAGAACGGGGCTCCCA..................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |