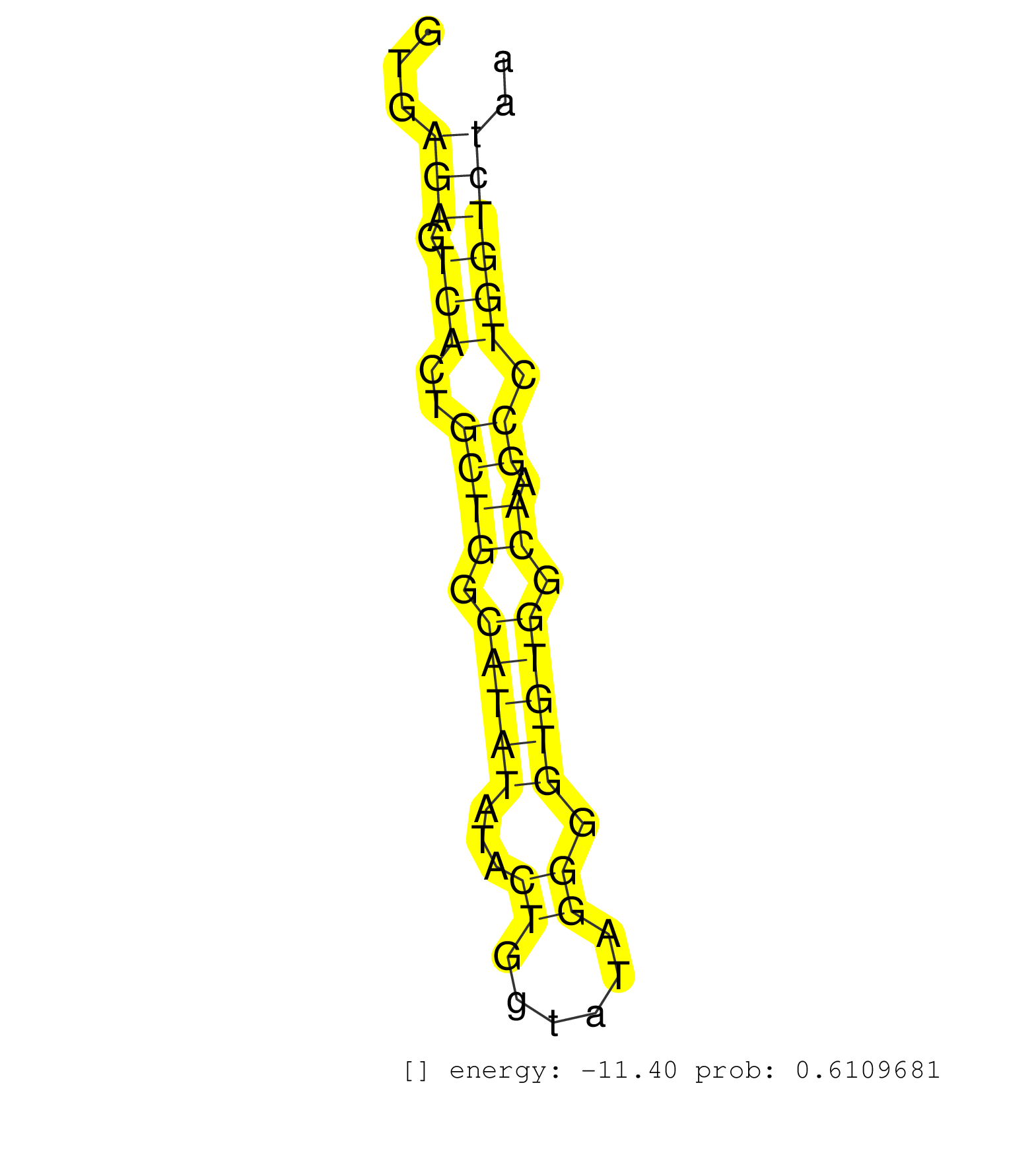

| Gene: Rusc2 | ID: uc008spm.1_intron_9_0_chr4_43439001_f | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(20) TESTES |
| CATCTCTTTGGTTCTCGGAAATCTCAGCGGGAAGCCCGGCCCACGAACAGGTGAGAGTCACTGCTGGCATATATACTGGTATAGGGGTGTGGCAAGCCTGGTCTAAATCAGGACACAGGCAGAACCAGGATCTGTCTGCTCTCCCCTAGGCTGCCGTCGGACTGGCTGAGCCTGGACAAGTCTGTGTTCCAGCTGGTAG .....................................................(((.(((..((((.(((((...((......)).))))).)).)).))))))............................................................................................... ..................................................51.....................................................106........................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGAGTCACTGCTGGCATATATACTG......................................................................................................................... | 28 | 1 | 12.00 | 12.00 | - | 4.00 | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - |
| ......TTTGGTTCTCGGAAATCTCAGCGG......................................................................................................................................................................... | 24 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGGGTGTGGCAAGCCTGGTga............................................................................................... | 23 | ga | 6.00 | 3.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GGCTGCCGTCGGACTGGCTG............................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGGGTGTGGCAAGCCTGGT................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................TAGGGGTGTGGCAAGCCTGG.................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GAGAGTCACTGCTGGCATATATACTG......................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TTTGGTTCTCGGAAATCTCAGCGGG........................................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ........TGGTTCTCGGAAATCTCAGCGGGAAGCC................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAAATCAGGACACAGGCAGAACCAGG...................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................TAAATCAGGACACAGGCAGAACCAGGtt.................................................................... | 28 | tt | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGGGTGTGGCAAGCCTGGTaat.............................................................................................. | 24 | aat | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............CTCGGAAATCTCAGCGGGAAGCCCGGCC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................ATAGGGGTGTGGCAAGCCTGGTt................................................................................................ | 23 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................TAGGGGTGTGGCAAGCCTGGTat............................................................................................... | 23 | at | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGTCACTGCTGGCATATATACT.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................ATAGGGGTGTGGCAAGCCTGGT................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....CTTTGGTTCTCGGAAATCTCAGCGGGAAGC.................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................TATATACTGGTATAGGGGTGTGGCA......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAAATCAGGACACAGGCAGAACCAG....................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........TGGTTCTCGGAAATCTCAGCGGGAAGC.................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................TAGGGGTGTGGCAAGCCTt................................................................................................... | 19 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TCTCAGCGGGAAGtag.................................................................................................................................................................. | 16 | tag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TCTCGGAAATCTCAGCGGGAAGCCCGGCC.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CAGGATCTGTCTGCTCTCCCCTAGt................................................. | 25 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGTCACTGCTGGCATATATACTGt........................................................................................................................ | 29 | t | 1.00 | 12.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGTCACTGCTGGCATATAaac........................................................................................................................... | 26 | aac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAGGGGTGTGGCAAGCCTGGTCa............................................................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAAATCAGGACACAGGCAGAACCAGGA..................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CATCTCTTTGGTTCTCGGAAATCTCAGCGGGAAGCCCGGCCCACGAACAGGTGAGAGTCACTGCTGGCATATATACTGGTATAGGGGTGTGGCAAGCCTGGTCTAAATCAGGACACAGGCAGAACCAGGATCTGTCTGCTCTCCCCTAGGCTGCCGTCGGACTGGCTGAGCCTGGACAAGTCTGTGTTCCAGCTGGTAG .....................................................(((.(((..((((.(((((...((......)).))))).)).)).))))))............................................................................................... ..................................................51.....................................................106........................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................CGTCGGACTGGCTGatca............................... | 18 | atca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |