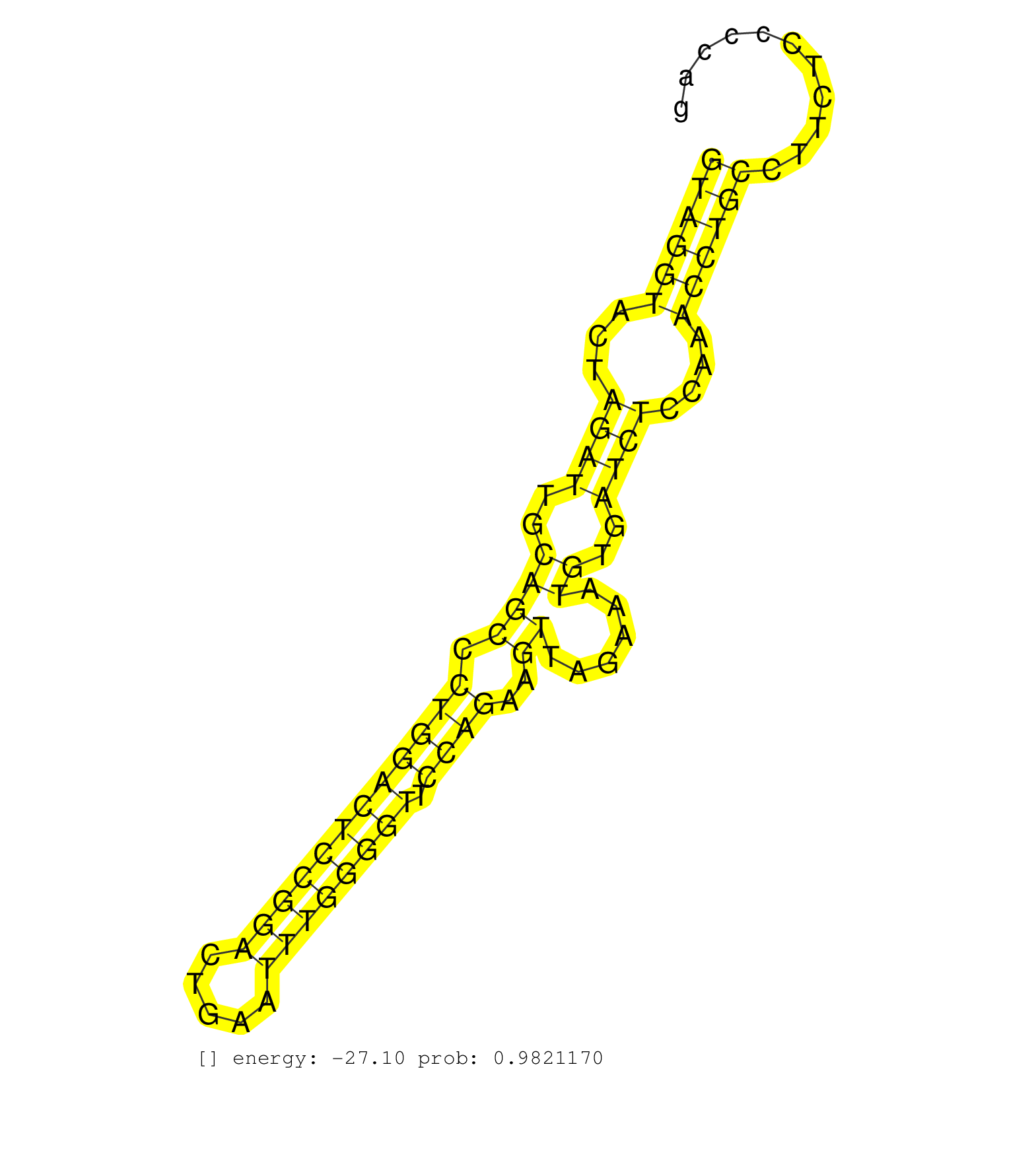
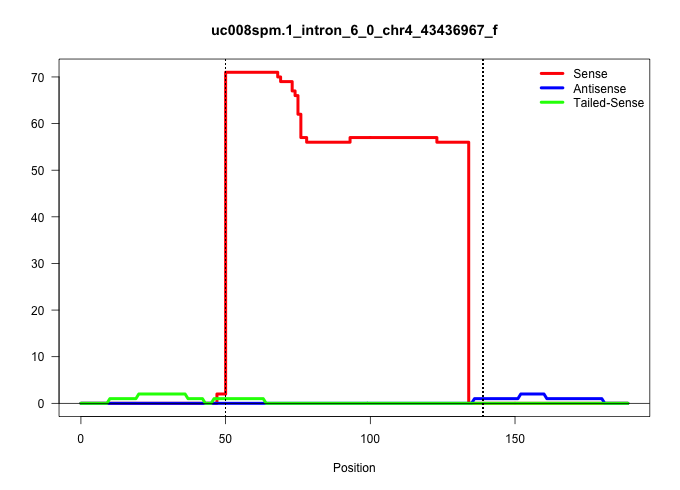
| Gene: Rusc2 | ID: uc008spm.1_intron_6_0_chr4_43436967_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| CCAACGTAAAAACATGCCTTGGAGTGTGGTCGAGGCTTCCACACAGCTAGGTAGGTACTAGATTGCAGCCCTGGACTCCGGACTGAATTTGGGGTTCCAGAAGTTAGAAATGTGATCTCCAAACCTGCCTTCTCCCCAGGCCCATCCACCAAGGTCTTGCATGGCCTCTACAACAAAGTCAGCCAGTTC ..................................................((((((...((((..((((.((((((((((((.....)))))))).))))..))......))..))))....))))))............................................................. ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTACTAGATTGCAGCCCTGGACTCCGGACTGAATTTGGGGTTCCAGAA....................................................................................... | 52 | 1 | 57.00 | 57.00 | 57.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTACTAGATTGCAGCCCTGGAC................................................................................................................. | 26 | 1 | 6.00 | 6.00 | - | - | 4.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGTACTAGATTGCAGCCCTGGA.................................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTAGGTACTAGATTGCAGCCCTG.................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTACTAGATTGCAG......................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTACTAGATTGCAGCCCTGGACTC............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GTTCCAGAAGTTAGAAATGTGATCTCCAAA.................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAGGTACTAGATTGCAGC........................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........AACATGCCTTGGAGTGTGGTCGAGGtt........................................................................................................................................................ | 27 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................TGGTCGAGGCTTCCACACAGCTA............................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................CTAGGTAGGTACTAGggc............................................................................................................................. | 18 | ggc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................GGAGTGTGGTCGAGGCTTCCACt.................................................................................................................................................. | 23 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGGTACTAGATTGCAGCCCTGG................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| CCAACGTAAAAACATGCCTTGGAGTGTGGTCGAGGCTTCCACACAGCTAGGTAGGTACTAGATTGCAGCCCTGGACTCCGGACTGAATTTGGGGTTCCAGAAGTTAGAAATGTGATCTCCAAACCTGCCTTCTCCCCAGGCCCATCCACCAAGGTCTTGCATGGCCTCTACAACAAAGTCAGCCAGTTC ..................................................((((((...((((..((((.((((((((((((.....)))))))).))))..))......))..))))....))))))............................................................. ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CAGGCCCATCCACCAAGGTCTTGCA............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................TCTTGCATGGCCTCTACAACAAAGTCAac........ | 29 | ac | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GGTCTTGCATGGCCTCTACAACAAAGTCA........ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |