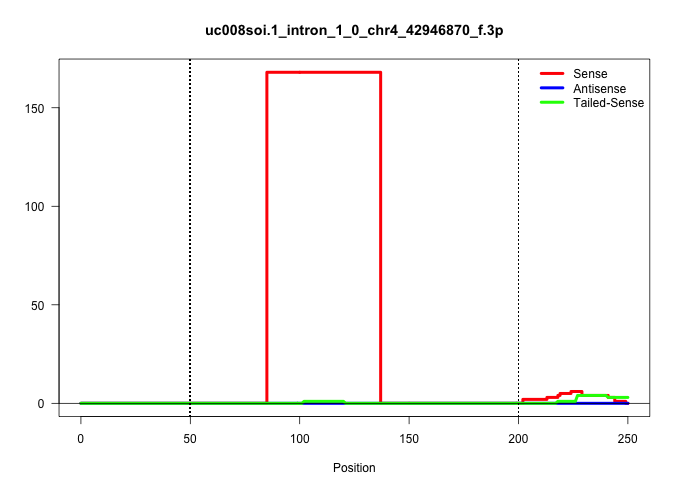
| Gene: N28178 | ID: uc008soi.1_intron_1_0_chr4_42946870_f.3p | SPECIES: mm9 |
 |
|
(2) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
| ATCACTCACACAGATTATTCTGTATAACATGGGAATAGAATCCCTGTTCAGAAACAGTAACAGAGCCCTAAGCAGCTTCCTAGTGCTCGTCCACCAGGAAGAACCCTTACACTTTAACTGTGGCCCTAGAGAGGTCTGGCCCATGCGGCAGGTGTGGGATCTTGCTATCTGTGCTTTCAAACCCTTCTCGTCCCTGCAAGGTTGTCAACGACGAGATGTGTGACGTCTGTGAAGTCTGGACCGCTGAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................CTCGTCCACCAGGAAGAACCCTTACACTTTAACTGTGGCCCTAGAGAGGTCT................................................................................................................. | 52 | 1 | 168.00 | 168.00 | 168.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTCAACGACGAGATGTGTGACGTCTG..................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................................TGTGAAGTCTGGACCGCTGAGAGccg | 26 | ccg | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGTGACGTCTGTGAAGTCTGGACCGC...... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GTGACGTCTGTGAAGTCTGGACCGC...... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................................GTCTGTGAAGTCTGGACCGCTGAGA. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TGTGAAGTCTGGACCGCTGAGAGcctt | 27 | cctt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGTGACGTCTGTGAAGTCTGGAt......... | 23 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................ACCCTTACACTTTAAgacg................................................................................................................................. | 19 | gacg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................AGATGTGTGACGTCTGTGAAGTCTGGAC......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ATCACTCACACAGATTATTCTGTATAACATGGGAATAGAATCCCTGTTCAGAAACAGTAACAGAGCCCTAAGCAGCTTCCTAGTGCTCGTCCACCAGGAAGAACCCTTACACTTTAACTGTGGCCCTAGAGAGGTCTGGCCCATGCGGCAGGTGTGGGATCTTGCTATCTGTGCTTTCAAACCCTTCTCGTCCCTGCAAGGTTGTCAACGACGAGATGTGTGACGTCTGTGAAGTCTGGACCGCTGAGAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................GATCTTGCTATCTGTtata.............................................................................. | 19 | tata | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |