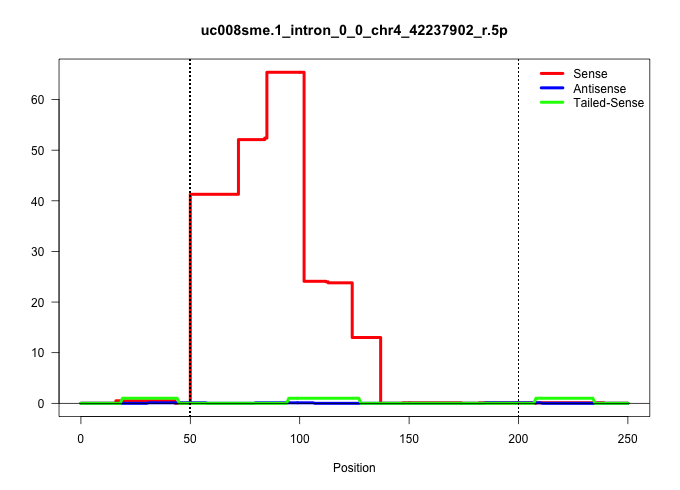
| Gene: AK006581 | ID: uc008sme.1_intron_0_0_chr4_42237902_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| AGTCCAGTCAGGAGGATGCAGAGGTTCACGGGCATCAGCCATGAGTCCAGGTAGACCAGTTGGACCAACCAACTCACTTCCCTATGCCTCAACTCTGTAGAGTCTCATTACCCTTTAGACAAAGGCCCTCAGTAGTGACCCCCCTGCTGCATGGGAGTGCCACTTGATTTCTTCCCCTTAAGCCCTCTTCTGTGACAACACTCCCTTCTAAAAACTCACCACTTGTTTCCCATGAAATCTGTTCTTCACG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGACCAGTTGGACCAACCAACTCACTTCCCTATGCCTCAACTCTGTAGAG.................................................................................................................................................... | 52 | 7 | 41.29 | 41.29 | 41.29 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GCCTCAACTCTGTAGAGTCTCATTACCCTTTAGACAAAGGCCCTCAGTAGTG................................................................................................................. | 52 | 2 | 13.00 | 13.00 | - | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CTCACTTCCCTATGCCTCAACTCTGTAGAGTCTCATTACCCTTTAGACAAAG.............................................................................................................................. | 52 | 10 | 10.80 | 10.80 | - | 10.80 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAAAACTCACCACTTGTTTCCCATGt............... | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................AGAGGTTCACGGGCATCAGCCATGAt............................................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGTAGAGTCTCATTACCCTTTAGACAAAGGCtt.......................................................................................................................... | 33 | tt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................TGCAGAGGTTCACGGGCATCAGCCATG............................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCAACTCTGTAGAGTCTCATTACCC......................................................................................................................................... | 29 | 10 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.10 | - | - | - | - | 0.10 | - |
| ................................................................................................................................................................................................................TAAAAACTCACCACTTGTTTCCCATGAAATC........... | 31 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - |
| ....................................................................................TGCCTCAACTCTGTAGAGTCTCATTACC.......................................................................................................................................... | 28 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - |
| ...................................................................................................................................................TGCATGGGAGTGCCACTTGATTTCTTC............................................................................ | 27 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - |
| ......................................................................................................................................................................................CCCTCTTCTGTGACAACACTCCCTT........................................... | 25 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| AGTCCAGTCAGGAGGATGCAGAGGTTCACGGGCATCAGCCATGAGTCCAGGTAGACCAGTTGGACCAACCAACTCACTTCCCTATGCCTCAACTCTGTAGAGTCTCATTACCCTTTAGACAAAGGCCCTCAGTAGTGACCCCCCTGCTGCATGGGAGTGCCACTTGATTTCTTCCCCTTAAGCCCTCTTCTGTGACAACACTCCCTTCTAAAAACTCACCACTTGTTTCCCATGAAATCTGTTCTTCACG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................CCAACTCACTTCCCagtg........................................................................................................................................................................ | 18 | agtg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TCTTCTGTGACAACACTCCCTTCTAA....................................... | 26 | 10 | 0.10 | 0.10 | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................CCTATGCCTCAACTCTGTAGAGTCTCA............................................................................................................................................... | 27 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - |
| ...............................GCATCAGCCATGAGTCCAGGTAGACCA................................................................................................................................................................................................ | 27 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - |