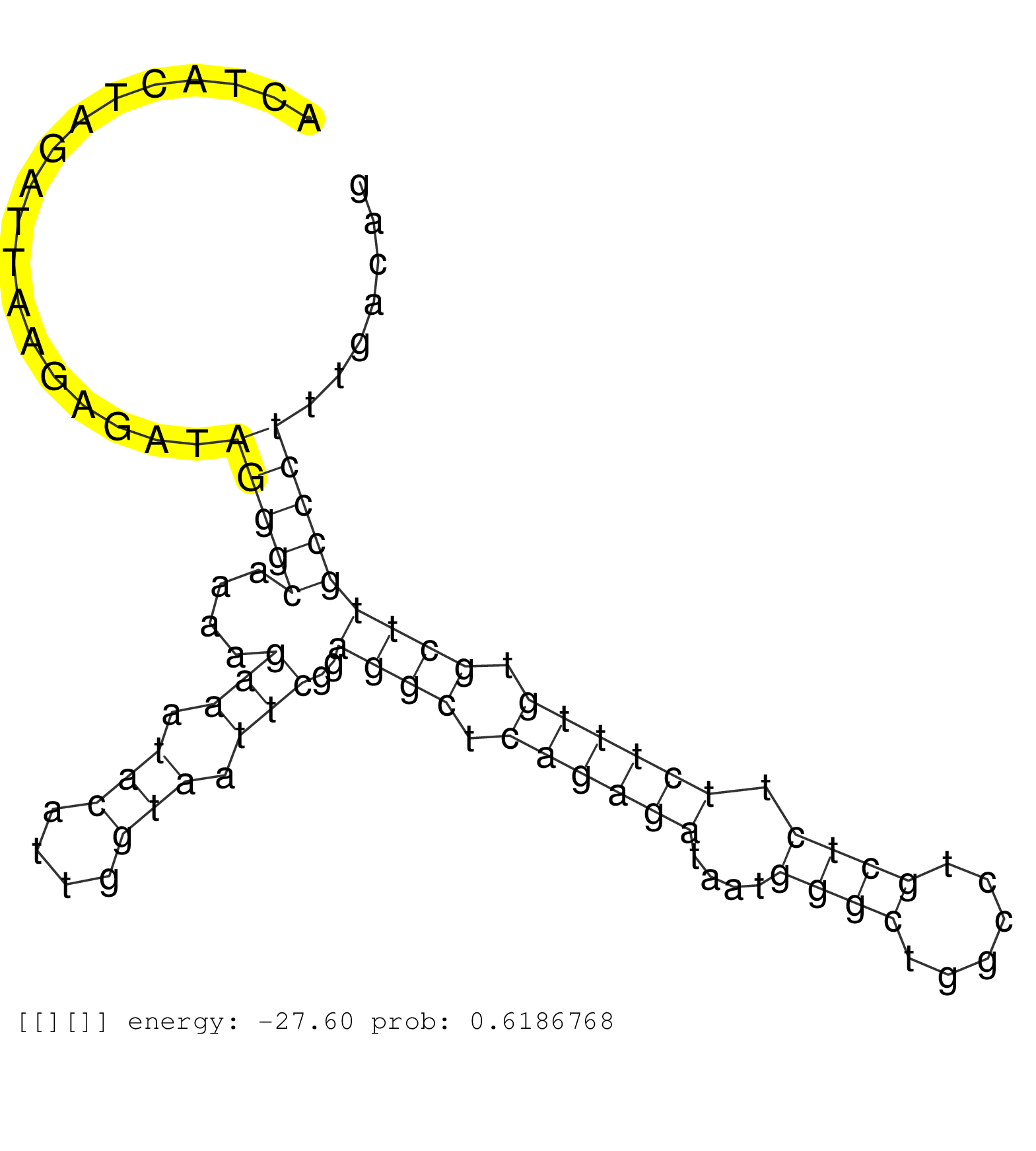

| Gene: Galt | ID: uc008sjn.1_intron_5_0_chr4_41703869_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| AAAATAAAGGAGCCATGATGGGCTGTTCTAACCCCCATCCCCACTGCCAGGCAAGTATGTGACAGGCCTTAATGGCTTTCTTGGCTGAGTCCACCGGCATTGTGGACAGGACTACTAGATTAAGAGATAGGGCAAAAGAAATACATTGGTAATTCGGAGGCTCAGAGATAATGGGCTGGCCTGCTCTTCTTTGTGCTTGCCCTTTGACAGGTTTGGGCTAGCAGCTTCCTGCCAGATATCGCCCAGCGTGAAGAGCGATC ................................................................................................................................(((((....(((.(((....))).)))..((((.((((((....((((......)))).)))))).)))))))))......................................................... ..............................................................................................................111................................................................................................210................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GTATGTGACAGGCCTTAATGGCTTTCTTGGCTGAGTCCACCGGCATTGTGGA.......................................................................................................................................................... | 52 | 1 | 126.00 | 126.00 | 126.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TGCCAGATATCGCCCAGCGTGAAGAGC.... | 27 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TGCCAGATATCGCCCAGCGTGAAGAGt.... | 27 | t | 3.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................................TGCCAGATATCGCCCAGCGTGAAGA...... | 25 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TAGCAGCTTCCTGCCAGATATCGCC................. | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TGCCAGATATCGCCCAGCGTGAAGAGCGA.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCTGCCAGATATCGCCCAGCGTGA......... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TGCCAGATATCGCCCAGCGTGAAGAGCa... | 28 | a | 1.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TAGCAGCTTCCTGCCAGATATCGCCCA............... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TAGCAGCTTCCTGCCAGATATCGCCC................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TTCCTGCCAGATATCGCCCAGCGTG.......... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................TTTGGGCTAGCAGCTTCCTGCCAGAT....................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GTTTGGGCTAGCAGCTTCCTGCCAGAT....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| AAAATAAAGGAGCCATGATGGGCTGTTCTAACCCCCATCCCCACTGCCAGGCAAGTATGTGACAGGCCTTAATGGCTTTCTTGGCTGAGTCCACCGGCATTGTGGACAGGACTACTAGATTAAGAGATAGGGCAAAAGAAATACATTGGTAATTCGGAGGCTCAGAGATAATGGGCTGGCCTGCTCTTCTTTGTGCTTGCCCTTTGACAGGTTTGGGCTAGCAGCTTCCTGCCAGATATCGCCCAGCGTGAAGAGCGATC ................................................................................................................................(((((....(((.(((....))).)))..((((.((((((....((((......)))).)))))).)))))))))......................................................... ..............................................................................................................111................................................................................................210................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................ACCGGCATTGTGGACAGGACTACTA............................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |