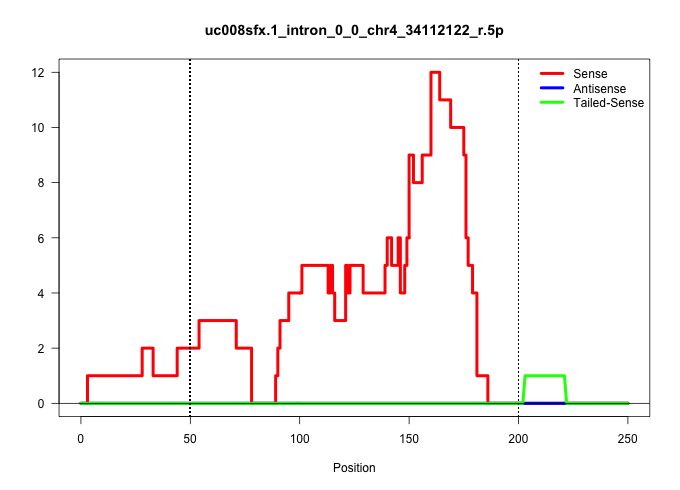
| Gene: Spaca1 | ID: uc008sfx.1_intron_0_0_chr4_34112122_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(16) TESTES |
| GGGTCAAAAACTTCAGCAACTGAGATACAGTCCGAGCTGAGCTCTATGAGGTATAAAGATTCGACGTCTCTCGACCAGTCACCAACAGATATACCTGTACATGAAGATGACGCTTTAAGTGAATGGAATGAATGATGTATGGATGACATATGAGAAACCAGAGACCATTAGAGCATATCAGATTCATTATTATCAAAATAAAATATATATTGGAAATTTTTCAATGATCTTGAATTGACTCTGCCTCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................GAGACCATTAGAGCATATCAG..................................................................... | 21 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AAAGATTCGACGTCTCTCGACCAG............................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AATGGAATGAATGATGTATGGATGA........................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGAATGAATGATGTATGGATGACATATG.................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................ACATATGAGAAACCAGAGACCATTAGAGCA........................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGAAGATGACGCTTTAAGTGAATGGAAT......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................ACCAGAGACCATTAGAGCATATCAGATTCA................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TTAAGTGAATGGAATGAATGATGTATGG............................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGAGAAACCAGAGACCATTAGAGCATATC....................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGATGACATATGAGAAACCAGAGACCATT................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TACCTGTACATGAAGATGACGC......................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGTACATGAAGATGACGCTTTAAGTGA................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................................TATATATTGGAAATTTaaa............................ | 19 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................TGAGAAACCAGAGACCATTAGAGCAT.......................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................ATACCTGTACATGAAGATGACGCTT....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................TATACCTGTACATGAAGATGACGCTTT...................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGAGAAACCAGAGACCATTAGAGCATA......................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGATGACATATGAGAAACCAGAGA...................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...TCAAAAACTTCAGCAACTGAGATACAGTCC......................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TATGAGGTATAAAGATTCGACGTCTCT................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................AGTCCGAGCTGAGCTCTATGAGGTAT.................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ATGAGAAACCAGAGACCATTAGAGCAT.......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TATGAGAAACCAGAGACCATTAGAGCAT.......................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGGTCAAAAACTTCAGCAACTGAGATACAGTCCGAGCTGAGCTCTATGAGGTATAAAGATTCGACGTCTCTCGACCAGTCACCAACAGATATACCTGTACATGAAGATGACGCTTTAAGTGAATGGAATGAATGATGTATGGATGACATATGAGAAACCAGAGACCATTAGAGCATATCAGATTCATTATTATCAAAATAAAATATATATTGGAAATTTTTCAATGATCTTGAATTGACTCTGCCTCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TTGGAAATTTTTCAAgtcc.......................... | 19 | gtcc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAAGATGACGCTTgcct....................................................................................................................................... | 18 | gcct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |