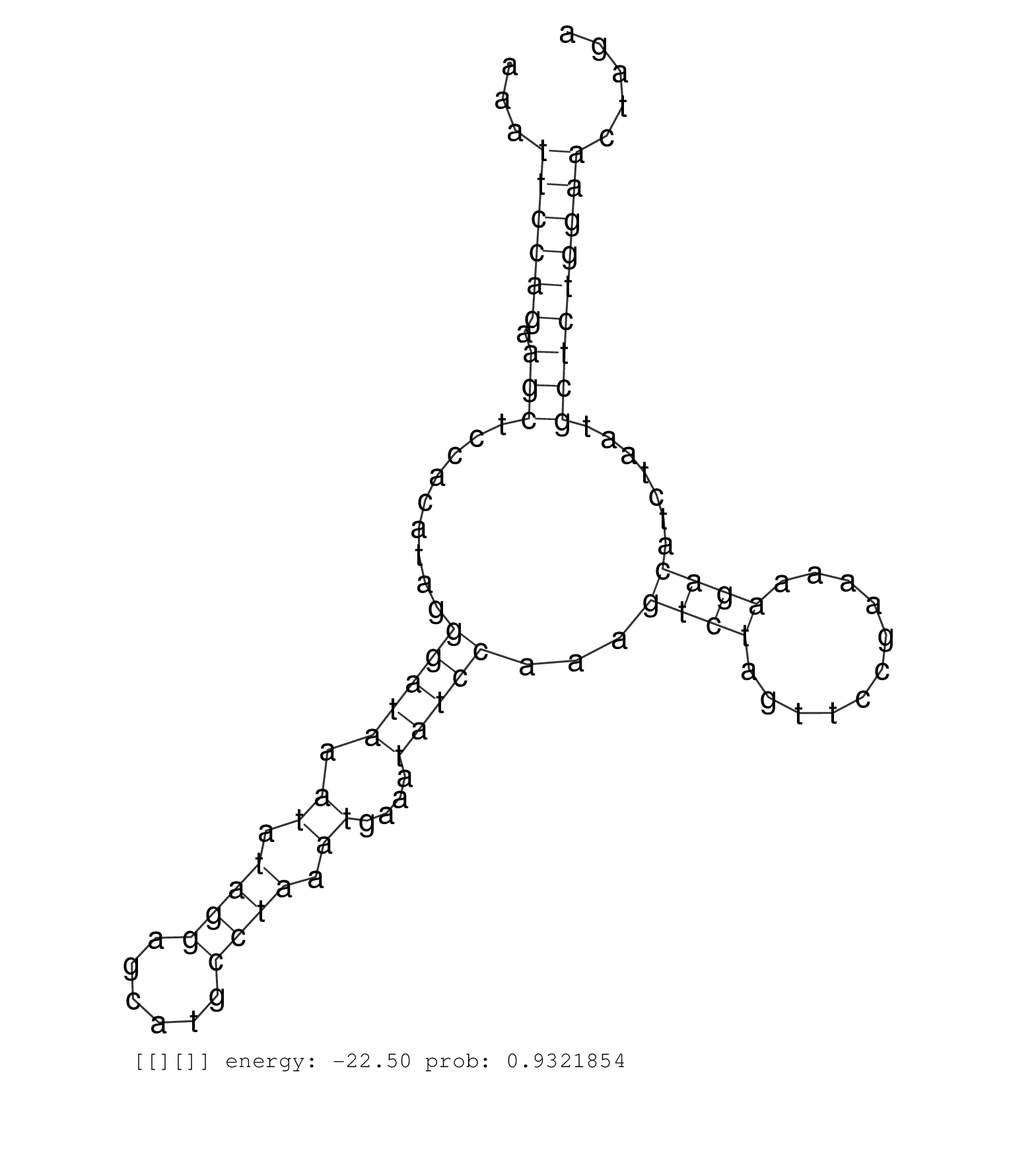
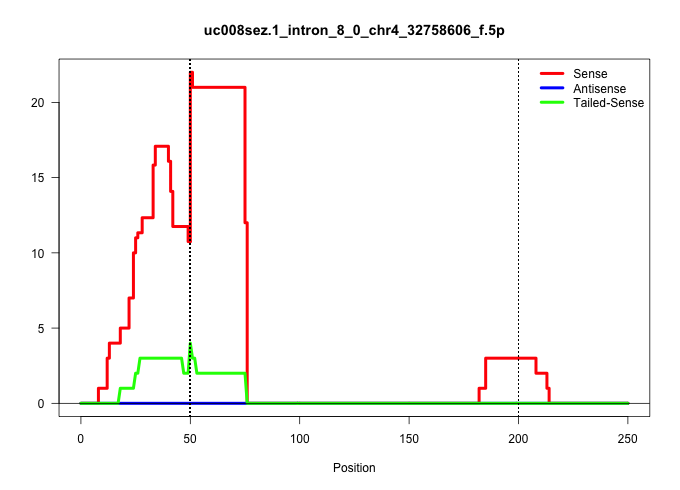
| Gene: Mdn1 | ID: uc008sez.1_intron_8_0_chr4_32758606_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| ACTGGACCAAAATTCACCTGCATAATCTGAATAAGAAAGACCTGAATGAGGTATGCAGCTTTGGATAATAAGAGGCTTTTGTTTCTATTTTTCATCATGAAAATTCCAGAAGCTCCACATAGGGATAAATATAGGAGCATGCCTAAATGAAATATCCAAAGTCTAGTTCCGAAAAAGACATCTAATGCTCTGGAACTAGAGATACTGCACTTTCAAACAGAATGTTGATCTCAGCACACCTAACAATTTT .......................................................................................................((((((.(((.........(((((.((.((((......)))).))....)))))...((((...........)))).......)))))))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGCAGCTTTGGATAATAAGAGGC.............................................................................................................................................................................. | 26 | 1 | 12.00 | 12.00 | 8.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGCAGCTTTGGATAATAAGAGG............................................................................................................................................................................... | 25 | 1 | 9.00 | 9.00 | 4.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| .................................AGAAAGACCTGAATGAG........................................................................................................................................................................................................ | 17 | 2 | 3.50 | 3.50 | 0.50 | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - |
| ......................TAATCTGAATAAGAAAGACCTGAATGAG........................................................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - |
| ........................ATCTGAATAAGAAAGACCTGAATGAG........................................................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................GAAAGACCTGAATGAG........................................................................................................................................................................................................ | 16 | 4 | 1.25 | 1.25 | - | - | - | - | - | 0.50 | 0.75 | - | - | - | - | - | - |
| ............TTCACCTGCATAATCTGAATAAGAAAGAC................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................GAATAAGAAAGACCTGAATGA......................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGCTCTGGAACTAGAGATACTGCACTTTC.................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............TTCACCTGCATAATCTGAATAAGAAAGACC................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTATGCAGCTTTGGATAATAAGAGGt.............................................................................................................................................................................. | 26 | t | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TCTGAATAAGAAAGACCTGAATGAGG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTATGCAGCTTTGGATAATAAGAGGa.............................................................................................................................................................................. | 26 | a | 1.00 | 9.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCATAATCTGAATAAGAAAGAC................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TAATGCTCTGGAACTAGAGATACTGC.......................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TCTGAATAAGAAAGACCTGAATGAGGTt..................................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................TGCATAATCTGAATAAGAAAGACCTGAAa........................................................................................................................................................................................................... | 29 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........AAAATTCACCTGCATAATCTGAATAAGAAAGA.................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............TCACCTGCATAATCTGAATAAGAAAGACC................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGCTCTGGAACTAGAGATACTGCACTTT..................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGAATAAGAAAGACCTGAATGAGa....................................................................................................................................................................................................... | 24 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CTGAATAAGAAAGACC................................................................................................................................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ACTGGACCAAAATTCACCTGCATAATCTGAATAAGAAAGACCTGAATGAGGTATGCAGCTTTGGATAATAAGAGGCTTTTGTTTCTATTTTTCATCATGAAAATTCCAGAAGCTCCACATAGGGATAAATATAGGAGCATGCCTAAATGAAATATCCAAAGTCTAGTTCCGAAAAAGACATCTAATGCTCTGGAACTAGAGATACTGCACTTTCAAACAGAATGTTGATCTCAGCACACCTAACAATTTT .......................................................................................................((((((.(((.........(((((.((.((((......)))).))....)))))...((((...........)))).......)))))))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|