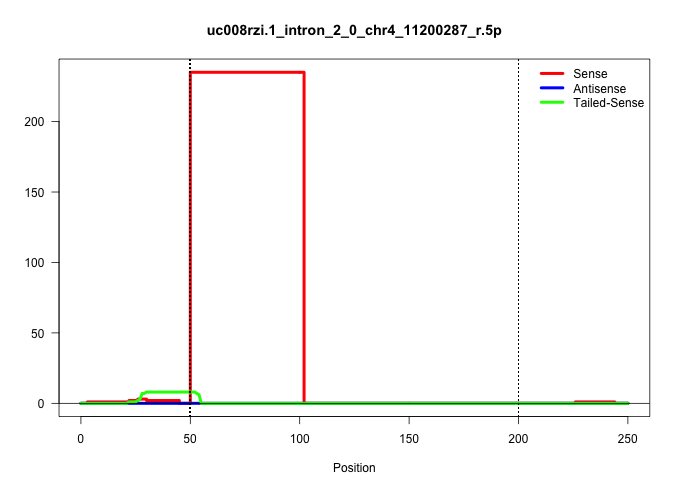
| Gene: Dpy19l4 | ID: uc008rzi.1_intron_2_0_chr4_11200287_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(9) TESTES |
| CTACAGGAATTCTATGATCCAGATACAGTGGAACTTATGACCTGGATAAAGTAAGGACCAAATTGCTCAAGACTATTGTTAACTGTCATTAACTAGTCTTTGCATGCTGTCTCATGACTTCGGGGATAATAATGAATGAATTGTTCTTTGTAAACTCAGGAATCATTAATTTGTAATTCTAATTTTATAAGTAAACTCATCTTAAAGTTGAAACAACCTTGGTAAGTCTGAGAGTGGCAGTTGGAGACCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGGACCAAATTGCTCAAGACTATTGTTAACTGTCATTAACTAGTCTTTG.................................................................................................................................................... | 52 | 1 | 235.00 | 235.00 | 235.00 | - | - | - | - | - | - | - | - |
| ............................TGGAACTTATGACCTGGATAAAGaggc................................................................................................................................................................................................... | 27 | aggc | 4.00 | 0.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - |
| ......................ATACAGTGGAACTTATGACCTGGATAAAGag..................................................................................................................................................................................................... | 31 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - |
| ..........................AGTGGAACTTATGACCTGGATAAAGagg.................................................................................................................................................................................................... | 28 | agg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...CAGGAATTCTATGATCCAGATACAGTG............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ......................ATACAGTGGAACTTATGACCTGG............................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..........................AGTGGAACTTATGACCTGG............................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCTGAGAGTGGCAGTTGG...... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ...........................GTGGAACTTATGACCTGGATAAAGaggt................................................................................................................................................................................................... | 28 | aggt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..............................GAACTTATGACCTGGATAAAGaggc................................................................................................................................................................................................... | 25 | aggc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| CTACAGGAATTCTATGATCCAGATACAGTGGAACTTATGACCTGGATAAAGTAAGGACCAAATTGCTCAAGACTATTGTTAACTGTCATTAACTAGTCTTTGCATGCTGTCTCATGACTTCGGGGATAATAATGAATGAATTGTTCTTTGTAAACTCAGGAATCATTAATTTGTAATTCTAATTTTATAAGTAAACTCATCTTAAAGTTGAAACAACCTTGGTAAGTCTGAGAGTGGCAGTTGGAGACCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|