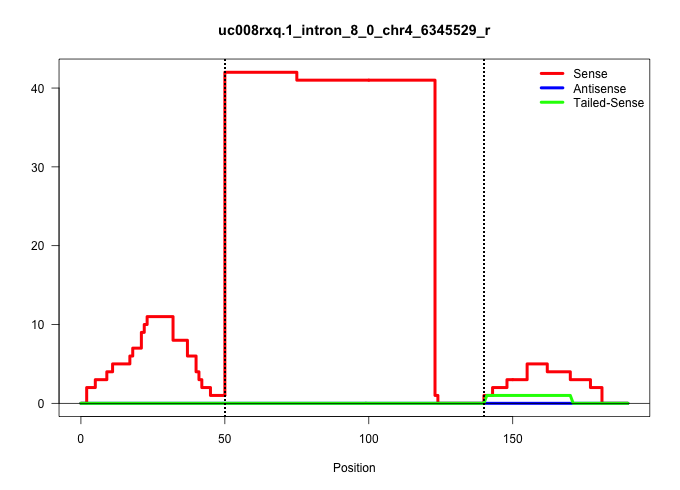
| Gene: Nsmaf | ID: uc008rxq.1_intron_8_0_chr4_6345529_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
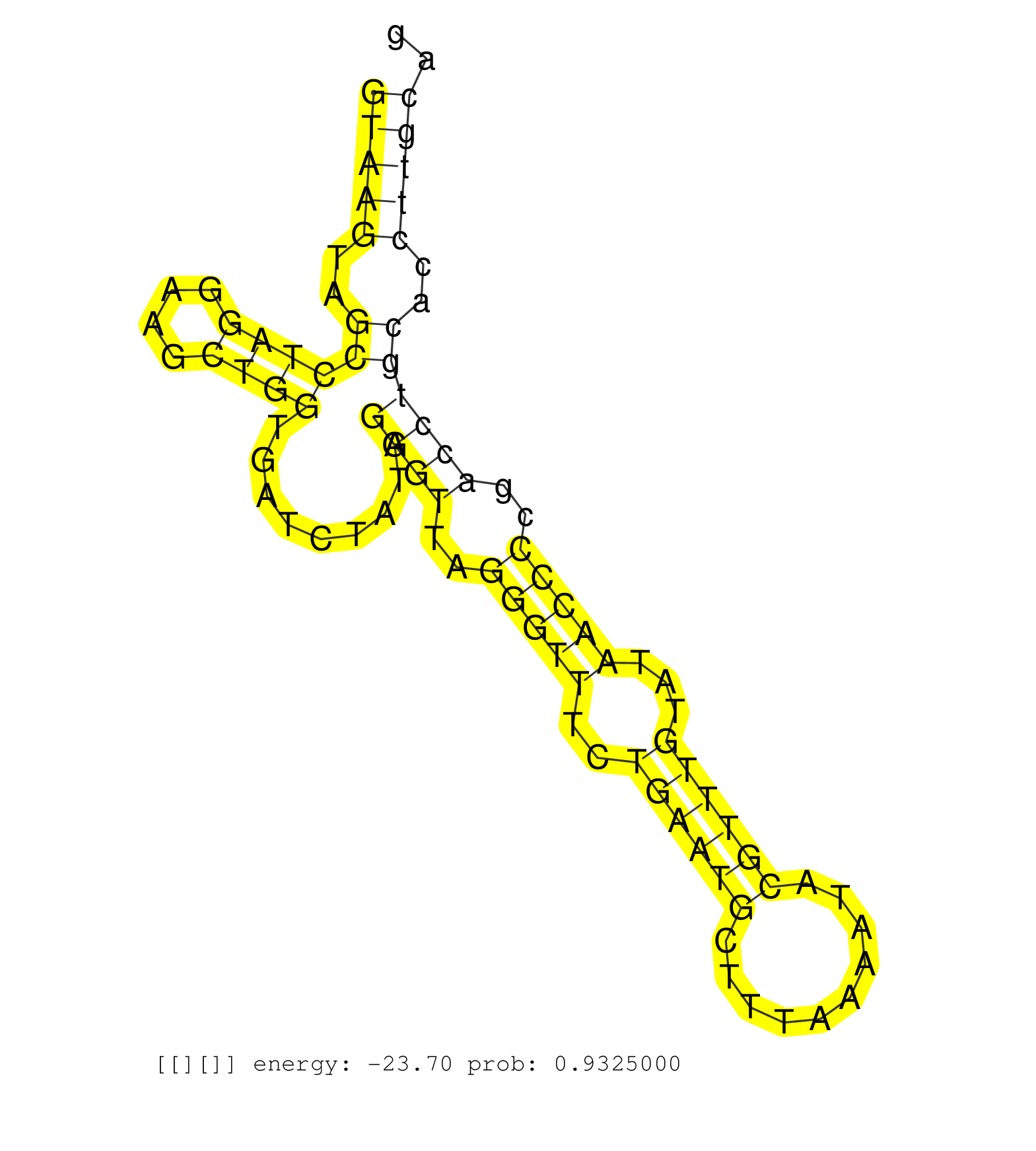
| TTGCAGAAACGTGGAAAAACTGTTTGGATGGTGCAACGGATTTTAAAGAGGTAAGTAGCCTAGGAAGCTGGTGATCTATAGGGTTAGGGTTTCTGAATGCTTTAAAATACGTTTGTATAACCCCGACCTGCACCTTGCAGTTGATTCCAGAATTCTATGATGAAGACGTCAGCTTTTTGGTCAATAGTCT ..................................................(((((..((((((....)))).........((((..(((((..((((((..........))))))...)))))..))))))..))))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTAGCCTAGGAAGCTGGTGATCTATAGGGTTAGGGTTTCTGAATGCTT........................................................................................ | 52 | 1 | 41.00 | 41.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GCAGAAACGTGGAAAAACTGTTTGGATGGT.............................................................................................................................................................. | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TATGATGAAGACGTCAGCTTTTTGGT......... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TTGATTCCAGAATTCTATGATGAAGACGTC.................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATTCCAGAATTCTATGATG............................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AATTCTATGATGAAGACGTCAGCTTTT............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................TGATTCCAGAATTCTATGATGAAGACGTCt................... | 30 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTTTGGATGGTGCAACGGA...................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCCAGAATTCTATGATGAAGACGTCA................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....GAAACGTGGAAAAACTGTTTGGATGGT.............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TTTGGATGGTGCAACGGATTTTA................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................AGAATTCTATGATGAAGACGTCAGCTTTT............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAGCCTAGGAAGCTGGTGAT................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........CGTGGAAAAACTGTTTGGATGGTGCAAC......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................GAATTCTATGATGAAGACGT..................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................AACTGTTTGGATGGTGCAACGGATT.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................GTTTGGATGGTGCAACGGAT..................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TTGGATGGTGCAACGGATTTTAAAGAG............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................ACTGTTTGGATGGTGCAACGGA...................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGAAAAACTGTTTGGATGGTGCAAC......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| TTGCAGAAACGTGGAAAAACTGTTTGGATGGTGCAACGGATTTTAAAGAGGTAAGTAGCCTAGGAAGCTGGTGATCTATAGGGTTAGGGTTTCTGAATGCTTTAAAATACGTTTGTATAACCCCGACCTGCACCTTGCAGTTGATTCCAGAATTCTATGATGAAGACGTCAGCTTTTTGGTCAATAGTCT ..................................................(((((..((((((....)))).........((((..(((((..((((((..........))))))...)))))..))))))..))))).................................................... ..................................................51.......................................................................................140................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TGCACCTTGCAGTTtcat................................................ | 18 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |