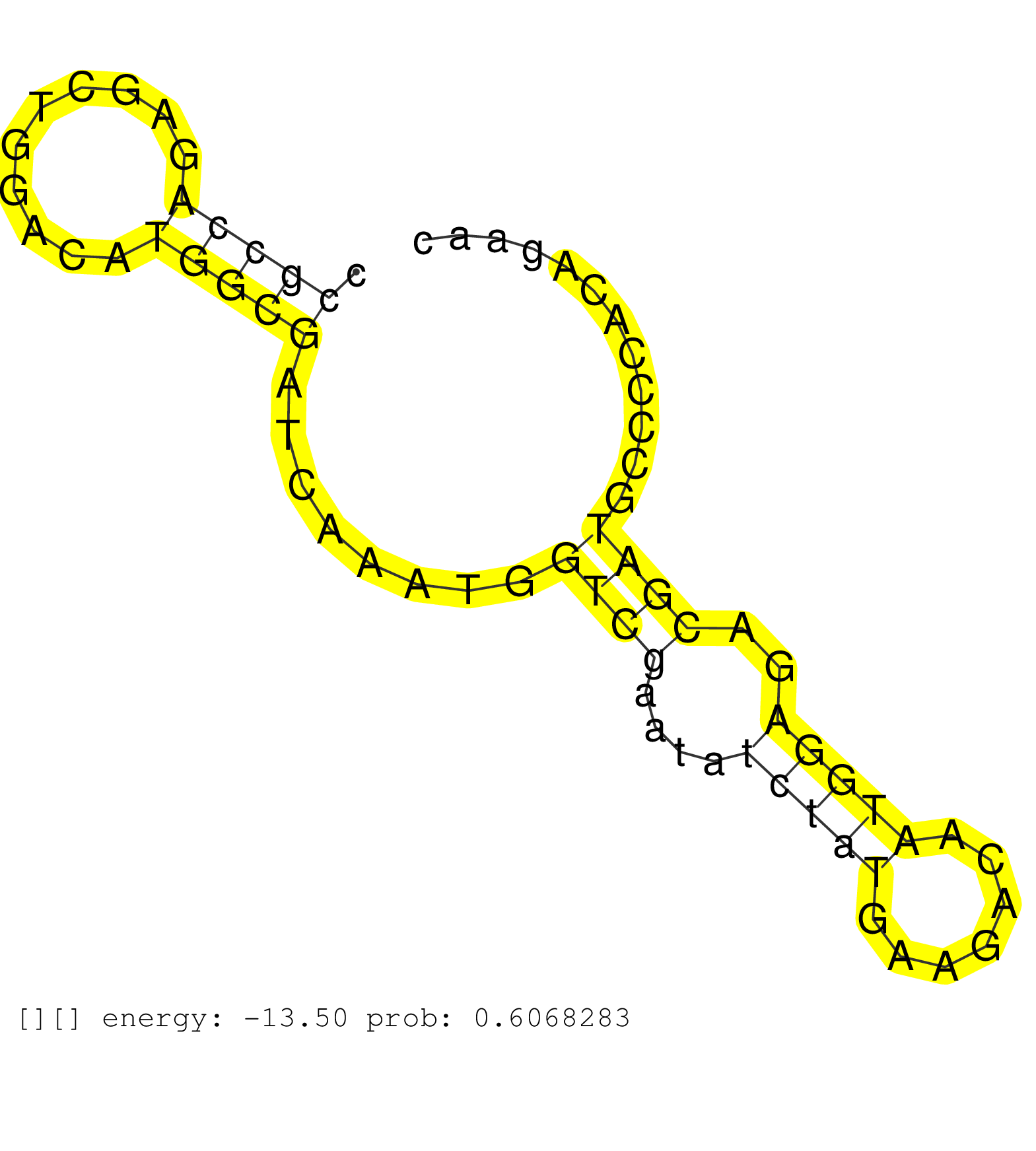

| Gene: 2410004B18Rik | ID: uc008rqu.1_intron_1_0_chr3_145601936_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| AAGTCCAACTGCGTGCCGCCCCCAGAGACCTACACTACTGAGAAGAAGCCGCCGCCGCCAGAGCTGGACATGGCGATCAAATGGTCGAATATCTATGAAGACAATGGAGACGATGCCCCACAGAACGCCAAGAAAGCCAGGCTTCTGCCTGAAGGGGAGGAGACAGTGGAATCAGGTAAGAGAGAG .......................................................(((((..........)))))........((((....(((((.......)))))..))))........................................................................ ......................................................55.....................................................................126.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................TGAAGACAATGGAGACGATGCCCCACA................................................................ | 27 | 1 | 24.00 | 24.00 | 7.00 | 4.00 | 3.00 | 4.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CGAATATCTATGAAGACAATGGAGACG.......................................................................... | 27 | 1 | 9.00 | 9.00 | 2.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATGCC..................................................................... | 28 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGAAGACAATGGAGACGATGCCCCAC................................................................. | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGGAGACGATGCCCCACAGAACGCCA........................................................ | 26 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GACCTACACTACTGAGAAGAAGCCGCC..................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CGAATATCTATGAAGACAATGGAGAC........................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATGCCCC................................................................... | 30 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCGAATATCTATGAAGACAATGGAGACG.......................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AGACCTACACTACTGAGAAGAAGCCGCC..................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCCTGAAGGGGAGGAGACAGTGGAA............... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCTATGAAGACAATGGAGACGATGCCC.................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATGCCC.................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGAGCTGGACATGGCGATCAAATGGTC.................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AATGGAGACGATGCCCCACAGAACGCCA........................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TATGAAGACAATGGAGACGATGCCCCACA................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GAAGACAATGGAGACGATGCCCCACA................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TACACTACTGAGAAGAAGCCGCCGCCG................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCCTGAAGGGGAGGAGACAGTGGAATC............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................TGGAGACGATGCCCCACAGAACGCCAA....................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGAAGACAATGGAGACGATGCCCCACt................................................................ | 27 | t | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGCGTGCCGCCCCCAGAGACCTACACTA..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATGCCCa................................................................... | 30 | a | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCCTGAAGGGGAGGAGACAGTGGA................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................GATCAAATGGTCGAATATCTATGAAG...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATG....................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGGAGACGATGCCCCACAGAACGCC......................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CGAATATCTATGAAGACAATGGAGACGt......................................................................... | 28 | t | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GAGACCTACACTACTGAGAAGAAGCCGCt..................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................AATGGTCGAATATCTATGAAGACAATG................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AAGACAATGGAGACGATGCCCCACAGAAC............................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGAAGACAATGGAGACGATGCCCCAaa................................................................ | 27 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................GAATATCTATGAAGACAATGGAGACG.......................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TACACTACTGAGAAGAAGCCGCCGCCGCCA.............................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGAGCTGGACATGGCGATCAAATGGTCGA.................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................ATCTATGAAGACAATGGAGACGATGCCCC................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TACACTACTGAGAAGAAGCCGCCGCCGCC............................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGTCGAATATCTATGAAGACAATGGAG............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................CAATGGAGACGATGCCCCACAGAACGCC......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................TATCTATGAAGACAATGGAGACGATGgc..................................................................... | 28 | gc | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GAGACCTACACTACTGAGAAGAAGCCG....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCCTGAAGGGGAGGAGACAGcgga................ | 25 | cgga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCTATGAAGACAATGGAGACGATGCCCCAC................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGAGACAGTGGAATCAG........... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| AAGTCCAACTGCGTGCCGCCCCCAGAGACCTACACTACTGAGAAGAAGCCGCCGCCGCCAGAGCTGGACATGGCGATCAAATGGTCGAATATCTATGAAGACAATGGAGACGATGCCCCACAGAACGCCAAGAAAGCCAGGCTTCTGCCTGAAGGGGAGGAGACAGTGGAATCAGGTAAGAGAGAG .......................................................(((((..........)))))........((((....(((((.......)))))..))))........................................................................ ......................................................55.....................................................................126.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................CATGGCGATCAAATGGTCGAA................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CAGGCTTCTGCCTGAAGGccat............................... | 22 | ccat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CAGGCTTCTGCCTGAAGc................................ | 18 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CAGGCTTCTGCCTGAAGt................................ | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................CAGGCTTCTGCCTGAAGcaa................................ | 20 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |