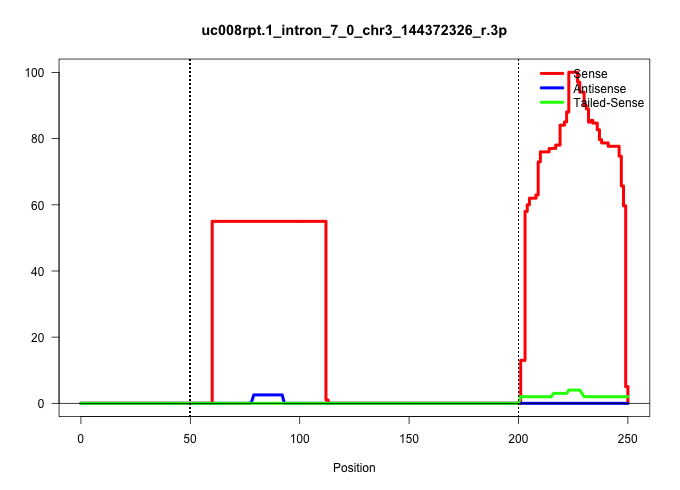
| Gene: Sh3glb1 | ID: uc008rpt.1_intron_7_0_chr3_144372326_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
| TACTTAAATAATTTTGCTAGTTTATTTAAAATTCTTTCCCCTTGTGGTTTGGGGAGTACAGGTCTTCGCTGTGCAGTGGACTCTAGCCTCGGATAACAGCATTTCCGCCTCCACCTCCCGTTGCTGAAGTTGCAGGCCTGTGCCACCATGCCTGCTTTTTCACTCTTTTGAAATACTGATATTCCTAACCTCTCTCCTAGATGCCAGGATAGAAGAATTTGTTTATGAGAAACTGGATAGAAAAGCACCA ............................................................................................................................(((((..((((((.((......)).))))))..)))))........................................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GGTCTTCGCTGTGCAGTGGACTCTAGCCTCGGATAACAGCATTTCCGCCTCC.......................................................................................................................................... | 52 | 1 | 55.00 | 55.00 | 55.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAG.......... | 17 | 1 | 10.00 | 10.00 | - | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAGAAAAGCACC. | 26 | 1 | 6.00 | 6.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAGAAAAGCACCA | 27 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTTTATGAGAAACTGGATAGAAAAGC.... | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATGAGA.................... | 29 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATGA...................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATG....................... | 26 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGTTTATGAGAAACTGGATAGAAAAGCAC.. | 29 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTATGAGAAACTGGATAGAAAAGCACC. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CCAGGATAGAAGAATTTGTTTATGAGAAA.................. | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTT.......................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAGA......... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATGCCAGGATAGAAGAATTTGTTTATGAG..................... | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AATTTGTTTATGAGAAACTGGATAG.......... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGAAGAATTTGTTTATGAGAAACTGG.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAGAAAAGCACCAt | 28 | t | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAGAAGAATTTGTTTATGAGAAACTGGA............. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ATGAGAAACTGGATAGAAAAGCA... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................................TAGAAGAATTTGTTTATGAGAAACTGG.............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATGAGt.................... | 29 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATGAt..................... | 28 | t | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTTGTTTATGAGAAACTGGA............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................TTATGAGAAACTGGATAGAAAAGCAC.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGAAGAATTTGTTTATGAGAAACTGGAT............ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TTTATGAGAAACTGGATAGAAAAGCAC.. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GCTGTGCAGTGGACTCTAG.................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGGATAGAAGAATTTGTTTATGAGAAA.................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAGGATAGAAGAATTTGTTTATGAGAAA.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGGATAGAAGAATTTGTTTATGAGAA................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTTGTTTATGAGAAACTGGATAGAAAAGCACC. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................AGAAGAATTTGTTTATGAGAAACTGGA............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GAATTTGTTTATGAGAAACTGGATAGA......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TATGAGAAACTGGATAGAAAAGCAC.. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................AAACTGGATAGAAAAGCACC. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAGGATAGAAGAATTTGTTTATGAGAAACT................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................................ATGAGAAACTGGATAGAAAAGCACC. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATAGAAGAATTTGTTTATGAGAAACTGGA............. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGCCAGGATAGAAGAATTTGTTTATGAG..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AAGAATTTGTTTATGAGAAACTGGATAGA......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................AGGATAGAAGAATTTGTTTATGA...................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TGGATAGAAAAGCACC. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................GGATAGAAAAGCACC. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................AATTTGTTTATGAGA.................... | 15 | 16 | 0.06 | 0.06 | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TACTTAAATAATTTTGCTAGTTTATTTAAAATTCTTTCCCCTTGTGGTTTGGGGAGTACAGGTCTTCGCTGTGCAGTGGACTCTAGCCTCGGATAACAGCATTTCCGCCTCCACCTCCCGTTGCTGAAGTTGCAGGCCTGTGCCACCATGCCTGCTTTTTCACTCTTTTGAAATACTGATATTCCTAACCTCTCTCCTAGATGCCAGGATAGAAGAATTTGTTTATGAGAAACTGGATAGAAAAGCACCA ............................................................................................................................(((((..((((((.((......)).))))))..)))))........................................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................GACTCTAGCCTCGGA............................................................................................................................................................. | 15 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | 0.20 | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - |