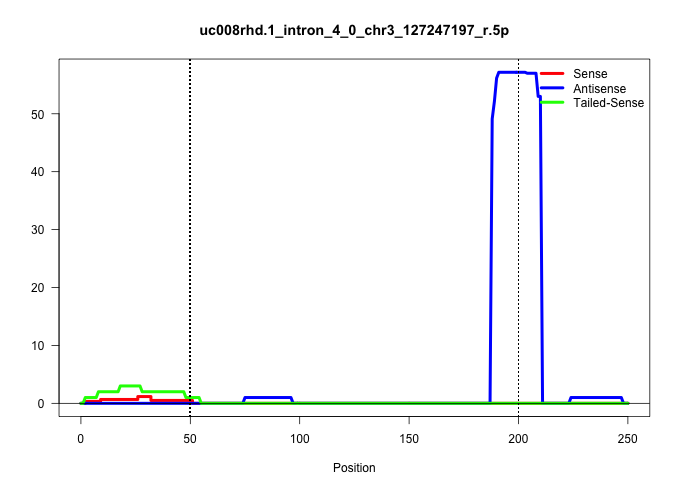
| Gene: Larp7 | ID: uc008rhd.1_intron_4_0_chr3_127247197_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| GAGAGACATAAGATGGGCGAGGAGGTTATACCATTGAGAGTGCTGTCCAAGTAAGTGGCCACTGACACAGGTCCATCACCATTGCTAAAGTGCAATTCCAATTTCTATTCAACAGAGTTGCATGTTAGCAACAGTGACGGCCTTGGTCTATCTATATGCAGCACAATGAAGATTAAGTAGGAGCCACCACACTCAAACATGGAAGCACTTATTTTTAAATGCACAGCAAGTGCCTCCATGTTAAAGTAAA ....................................................................................................................(((((......))))).(((..(((((((....((((.(((........))).))))....)))).)))..)))............................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................TGAGAGTGCTGTCCAAGactt................................................................................................................................................................................................... | 21 | actt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........TAAGATGGGCGAGGAGGTTATACCAa........................................................................................................................................................................................................................ | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GAGGAGGTTATACCATTGAGAGTGCTGTCt.......................................................................................................................................................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..GAGACATAAGATGGGCGAGGAGGTTt.............................................................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................TATACCATTGAGAGTGCTGTCCAAG....................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..GAGACATAAGATGGGCGAGGAGGTTATACC.......................................................................................................................................................................................................................... | 30 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .........AAGATGGGCGAGGAGGTTATACC.......................................................................................................................................................................................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GAGAGACATAAGATGGGCGAGGAGGTTATACCATTGAGAGTGCTGTCCAAGTAAGTGGCCACTGACACAGGTCCATCACCATTGCTAAAGTGCAATTCCAATTTCTATTCAACAGAGTTGCATGTTAGCAACAGTGACGGCCTTGGTCTATCTATATGCAGCACAATGAAGATTAAGTAGGAGCCACCACACTCAAACATGGAAGCACTTATTTTTAAATGCACAGCAAGTGCCTCCATGTTAAAGTAAA ....................................................................................................................(((((......))))).(((..(((((((....((((.(((........))).))))....)))).)))..)))............................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................ACACTCAAACATGGAAGCACTTA....................................... | 23 | 1 | 50.00 | 50.00 | 15.00 | 17.00 | 7.00 | - | 1.00 | 3.00 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................ACTCAAACATGGAAGCACTTA....................................... | 21 | 1 | 5.00 | 5.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCACCATTGCTAAAGTGCAAT.......................................................................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................ACACTCAAACATGGAAGCACT......................................... | 21 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................CACTCAAACATGGAAGCACTTA....................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................CTCAAACATGGAAGCACTTA....................................... | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................AAACATGGAAGCACTTAagt....................................... | 20 | agt | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCACCATTGCTAAAGTGCAATT......................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................ACACTCAAACATGGAAGCACTTAt....................................... | 24 | t | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TCACCATTGCTAAAGTGCAATgt.......................................................................................................................................................... | 23 | gt | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................AAACATGGAAGCACTTAtgga....................................... | 21 | tgga | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................CACTCAAACATGGAAGCACTTAa....................................... | 23 | a | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................ACCATTGCTAAAGTGCAAT.......................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................CTCAAACATGGAAGCACTTAg....................................... | 21 | g | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TCAAACATGGAAGCACTTAatgt....................................... | 23 | atgt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................AAACATGGAAGCACTTAtaga....................................... | 21 | taga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................AGCAAGTGCCTCCATGTTAAAGTA.. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TCAAACATGGAAGCACTTA....................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TCACCATTGCTAAAGTGCAATTa......................................................................................................................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................ACACTCAAACATGGAA.............................................. | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - |