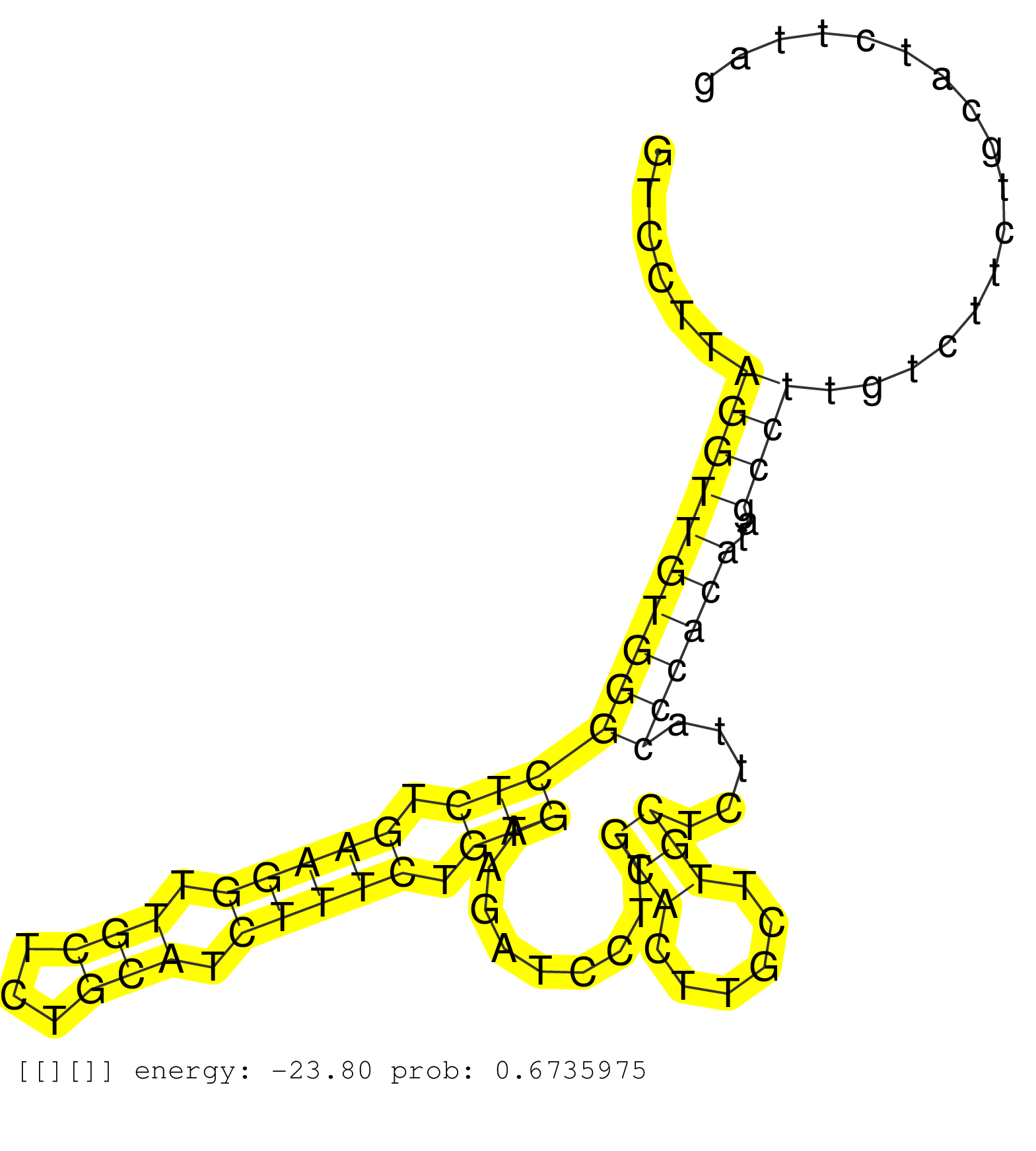
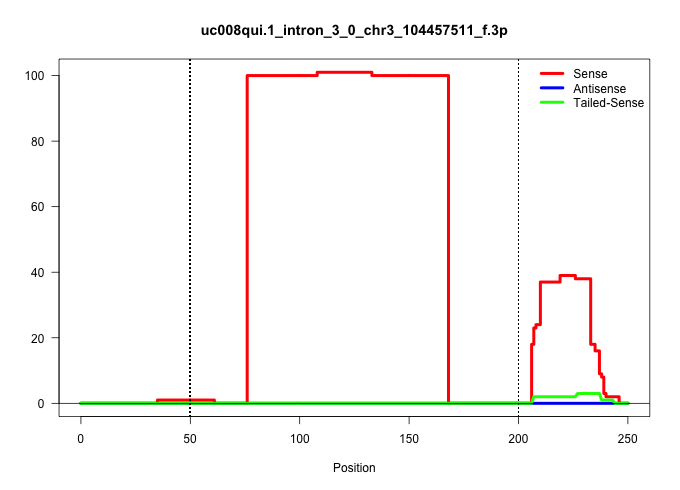
| Gene: Slc16a1 | ID: uc008qui.1_intron_3_0_chr3_104457511_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(14) TESTES |
| GATTTGCCAAGAGTTAATTGTTGCCAATTTGAAAATAGAAAATACAAGGCCAGTCAAAATCAGGAACACTATAAATAGAAAATCTTGCAGTTCATTTTTGGTCCTTAGGTTGTGGGCTCTGAAGGTTGCTCTGCATCTTTCTGAGTAGATCCTTGCACTTGCTTGCTCTTACCCACATAGCCTTGTCTTCTGCATCTTAGGCCGCCTCAATGACATGTATGGAGACTACAAATACACGTACTGGGCTTGT ..........................................................................................................(((((((((((((.(((((.(((...))).))))).))).........(((......))).....))))))..))))................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................AGAAAATCTTGCAGTTCATTTTTGGTCCTTAGGTTGTGGGCTCTGAAGGTTG.......................................................................................................................... | 52 | 1 | 100.00 | 100.00 | 100.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCAATGACATGTATGGAGACTACAAAT................. | 27 | 1 | 17.00 | 17.00 | - | 4.00 | 5.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGACATGTATGGAGACTACAAATACAC............. | 27 | 1 | 6.00 | 6.00 | - | 2.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGACATGTATGGAGACTACAAATACACGT........... | 29 | 1 | 5.00 | 5.00 | - | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CAATGACATGTATGGAGACTACAAAT................. | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................CAATGACATGTATGGAGACTACAAATACACt............ | 31 | t | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGGAGACTACAAATACACGTACTGGGC.... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ACAAATACACGTACTGt...... | 17 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................GTTGTGGGCTCTGAAGGTTGCTCTG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCAATGACATGTATGGAGACTACAAATAC............... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGACATGTATGGAGACTACAAATACACG............ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GGTCCTTAGGTTGTGGGCTCTGAAG.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................CAATGACATGTATGGAGACTACAAATAC............... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CAATGACATGTATGGAGACTACAAATACAC............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................TAGAAAATACAAGGCCAGTCAAAATC............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGACATGTATGGAGACTACAAATACACGTA.......... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AATGACATGTATGGAGAC........................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GATTTGCCAAGAGTTAATTGTTGCCAATTTGAAAATAGAAAATACAAGGCCAGTCAAAATCAGGAACACTATAAATAGAAAATCTTGCAGTTCATTTTTGGTCCTTAGGTTGTGGGCTCTGAAGGTTGCTCTGCATCTTTCTGAGTAGATCCTTGCACTTGCTTGCTCTTACCCACATAGCCTTGTCTTCTGCATCTTAGGCCGCCTCAATGACATGTATGGAGACTACAAATACACGTACTGGGCTTGT ..........................................................................................................(((((((((((((.(((((.(((...))).))))).))).........(((......))).....))))))..))))................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TCCTTGCACTTGCTcctt....................................................................................... | 18 | cctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |