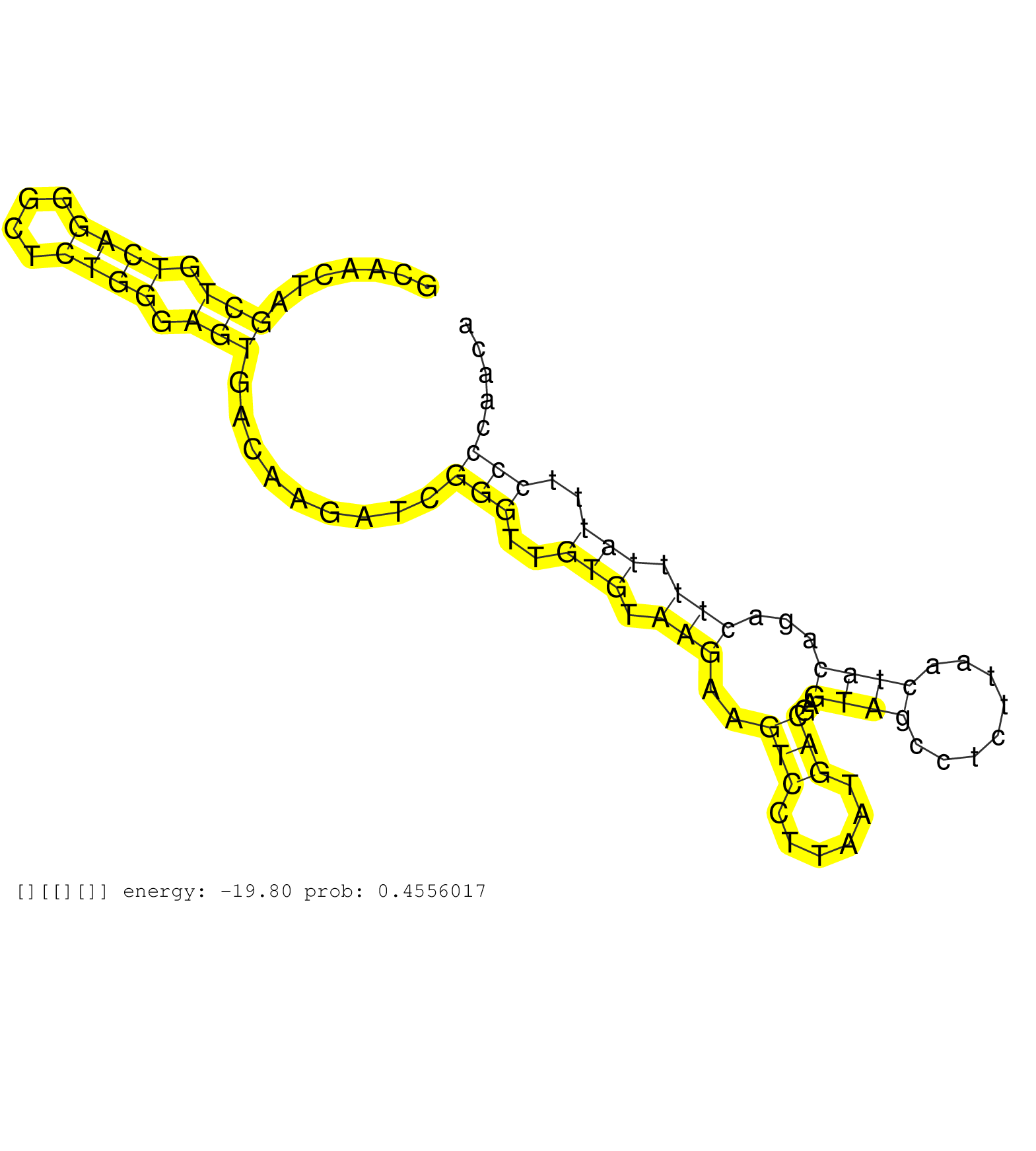

| Gene: Olfml3 | ID: uc008qtf.1_intron_1_0_chr3_103541161_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(10) TESTES |
| AGCAGCACCACCTTGTGGAGTACATGGAACGCCGACTAGCTGCCTTAGAGGTGAGGGACTTGTTTCTCTTCTCACCCAGGATGAGTCCACATCTATAGGTGCAACTAGCTGTCAGGGCTCTGGGAGTGACAAGATCGGGTTGTGTAAGAAGTCCTTAATGACGAGTAGCCTCTTAACTACAGACTTTTATTTCCCCAACATAAGACAATTGCCCAGCAAATAAACGCAACACGGCTCCCTCAGATTAGAT ...........................................................................................................(((.((((....)))).))).........(((..(((.(((..(((......)))..((((........))))...))).)))..)))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................ATAGGTGCAACTAGCTGTCAGGGCTCTGGGAGTGACAAGATCGGGTTGTGTA........................................................................................................ | 52 | 1 | 51.00 | 51.00 | 51.00 | - | - | - | - | - | - | - | - | - |
| .............TGTGGAGTACATGGAACGCCGACTAGCTGC............................................................................................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TAAGACAATTGCCCggtc................................ | 18 | ggtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................TATAGGTGCAACTAGCTGTCAGGGCTC.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................TAAGACAATTGCCCAGCAAATAAACGa....................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................GCCTTAGAGGTGAGGGACTTGTTTC........................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TAGCTGTCAGGGCTCTGGGAGT........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................TGACAAGATCGGGTTGTGTAAGA..................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............TGGAGTACATGGAACGCCGACTAGCTGC............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| AGCAGCACCACCTTGTGGAGTACATGGAACGCCGACTAGCTGCCTTAGAGGTGAGGGACTTGTTTCTCTTCTCACCCAGGATGAGTCCACATCTATAGGTGCAACTAGCTGTCAGGGCTCTGGGAGTGACAAGATCGGGTTGTGTAAGAAGTCCTTAATGACGAGTAGCCTCTTAACTACAGACTTTTATTTCCCCAACATAAGACAATTGCCCAGCAAATAAACGCAACACGGCTCCCTCAGATTAGAT ...........................................................................................................(((.((((....)))).))).........(((..(((.(((..(((......)))..((((........))))...))).)))..)))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................CCTCTTAACTACAGACTTTTATTTCCCC...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................GTGCAACTAGCTGTgtgg.......................................................................................................................................... | 18 | gtgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................TCTCACCCAGGATGAtctc...................................................................................................................................................................... | 19 | tctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |