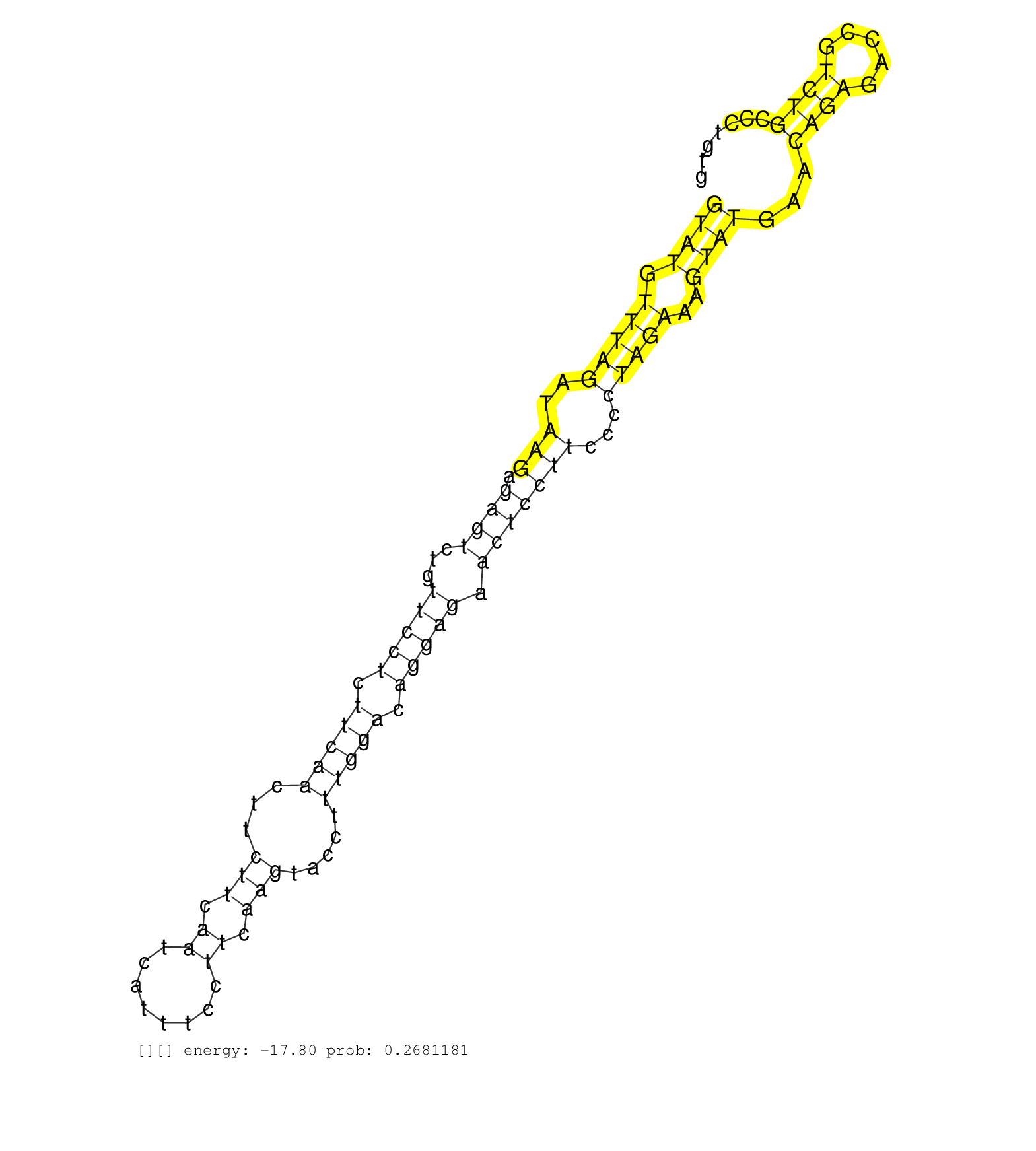

| Gene: Polr3c | ID: uc008qoa.1_intron_12_0_chr3_96527976_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(7) PIWI.ip |
(3) PIWI.mut |
(23) TESTES |
| GCCAGCCACTTAGAGTAATTGCCCACGACACAAAAGCATCCCTGGACCAGGTATGTTTAGATAAGAGAGTCTGTTCCTCTTCAACTTCTTCAATCATTTCCTTCAAGTACCTTTGGACAGGAGAACTCCTTCCCCTAGAAAGTATGAACAGAGACCGTCTGCCCTGTGGTCAGTACCACAGTGGGTGGGGTGGGGTGTGGTATTGTACTTAGGCCCCGTGTACTGCCGCTTGTGGTGTTCCTAGCTCATC ..................................................((((.(((((..(((.((((...(((((.(((((...(((.((........)).))).....))))).))))).)))))))...)))))..))))...((((.....))))......................................................................................... ..................................................51...................................................................................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCTGCCC...................................................................................... | 29 | 1 | 61.00 | 61.00 | 21.00 | 20.00 | 1.00 | 6.00 | 1.00 | - | 7.00 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCTGC........................................................................................ | 27 | 1 | 27.00 | 27.00 | 7.00 | - | 7.00 | 1.00 | 6.00 | - | - | - | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCTGCC....................................................................................... | 28 | 1 | 12.00 | 12.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCTG......................................................................................... | 26 | 1 | 8.00 | 8.00 | 2.00 | 2.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TTAGAGTAATTGCCCACGACACAAAAGC..................................................................................................................................................................................................................... | 28 | 1 | 7.00 | 7.00 | - | - | - | - | - | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCTGCCCT..................................................................................... | 30 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TTAGAGTAATTGCCCACGACACAAAAG...................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TGCCCACGACACAAAAGCATCCCTGGAC........................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............GAGTAATTGCCCACGACACAAAAGCAT................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........TAGAGTAATTGCCCACGACACAAAAGCA.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............GAGTAATTGCCCACGACACAAAAGCATCC................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CCAGGTATGTTTAGAagtt......................................................................................................................................................................................... | 19 | agtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................TGGTATTGTACTTAGGCCCCGTGTAaa.......................... | 27 | aa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............GAGTAATTGCCCACGACACAAAAGCATCCC................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGACAGGAGAACTCCTTCCCC................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAAAGTATGAACAGAGACCGTCTGCCC...................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CCCCTAGAAAGTATGAACAGAGACCGT............................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CGACACAAAAGCATCCCTGGACCAGGT...................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............GAGTAATTGCCCACGACACAAAAGCA.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TTAGAGTAATTGCCCACGACACAAAcg...................................................................................................................................................................................................................... | 27 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTTAGAGTAATTGCCCACGACACAAAAGC..................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTATGTTTAGATAAGAGAGTCTGTTC.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGA................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................ACGACACAAAAGCATCCCTGGACCAGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TAGAGTAATTGCCCACGACACAAAAGCATC.................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AACAGAGACCGTCTGCCCTGTGGTCAGT............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TAGAAAGTATGAACAGAGACCGTCT.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCTTCCCCTAGAAAGTATGAACAGAGACCGTC........................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CACTTAGAGTAATTGCCCACGACAC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TACCTTTGGACAGGttct............................................................................................................................. | 18 | ttct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................TTGTACTTAGGCCCCGTGTACTGCCG...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGGACCAGGTATGTTTAGATAAGAGAGTC................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCAGCCACTTAGAGTAATTGCCCACGACACAAAAGCATCCCTGGACCAGGTATGTTTAGATAAGAGAGTCTGTTCCTCTTCAACTTCTTCAATCATTTCCTTCAAGTACCTTTGGACAGGAGAACTCCTTCCCCTAGAAAGTATGAACAGAGACCGTCTGCCCTGTGGTCAGTACCACAGTGGGTGGGGTGGGGTGTGGTATTGTACTTAGGCCCCGTGTACTGCCGCTTGTGGTGTTCCTAGCTCATC ..................................................((((.(((((..(((.((((...(((((.(((((...(((.((........)).))).....))))).))))).)))))))...)))))..))))...((((.....))))......................................................................................... ..................................................51...................................................................................................................168................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|