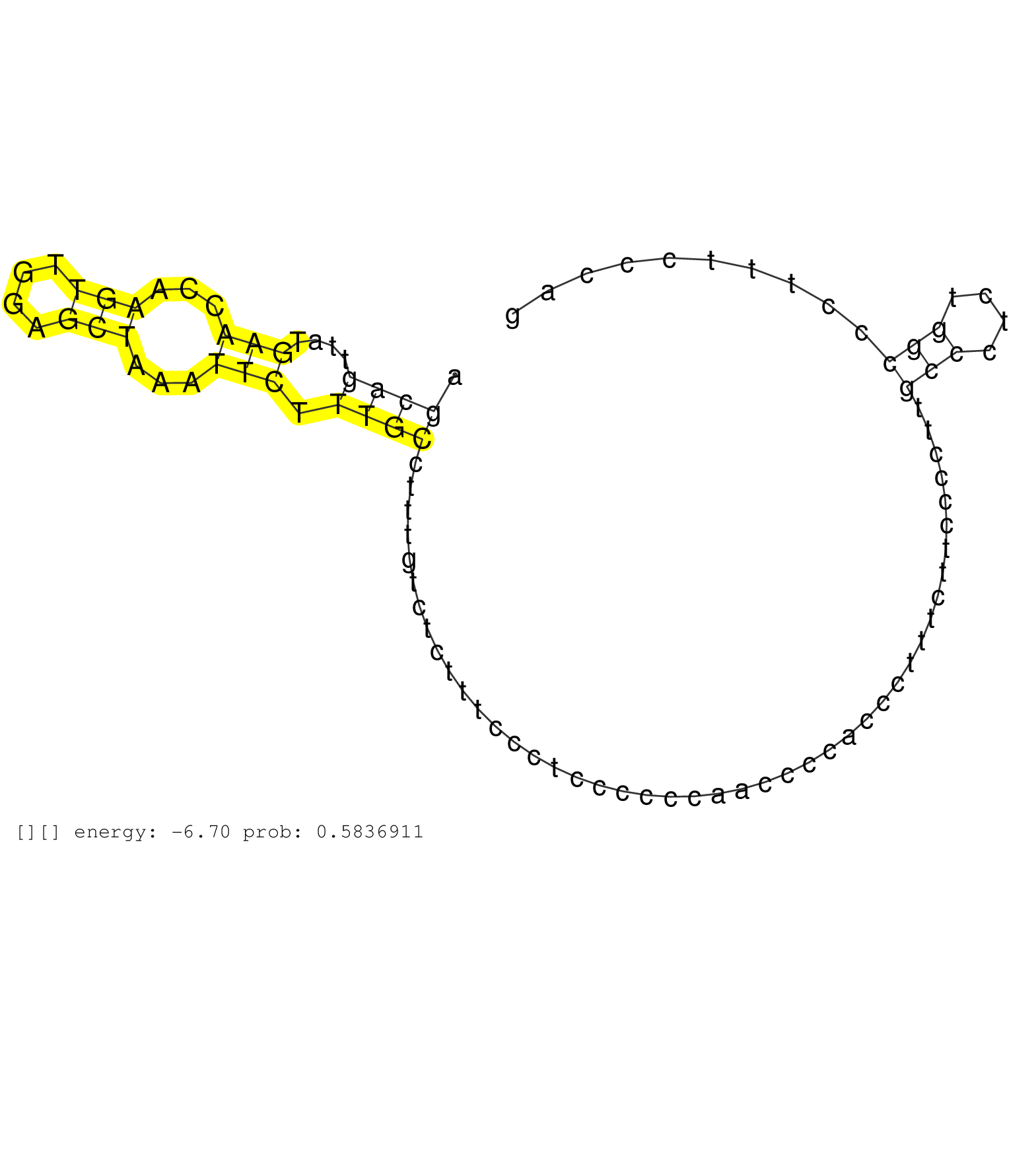
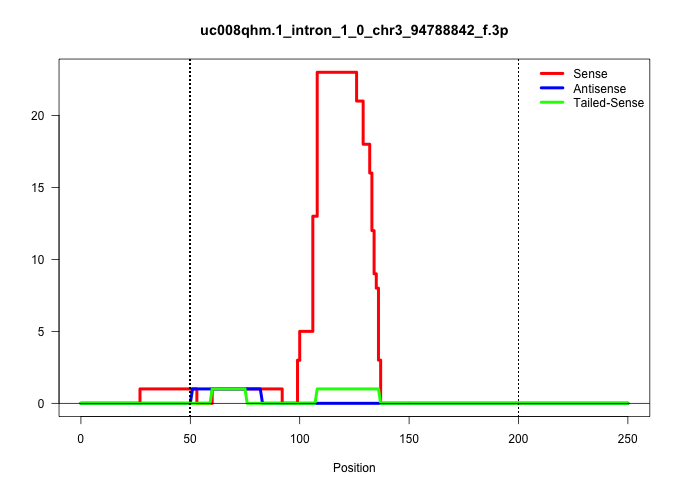
| Gene: Pi4kb | ID: uc008qhm.1_intron_1_0_chr3_94788842_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(12) TESTES |
| TGAGCTGGCCTTCCAACATCCCACAGATGTCTAGACTCTGGGCAACTGGCAGTTTCAGGCTGACTGTCTTTCACTAAAGCAGATATAGCTGCACAAGAGAAGCAGTTATGAACCAAGTTGGAGCTAAATTCTTTGCCTTTGTCTCTTTCCCTCCCCCCAACCCCACCCTTTCTTCCCCTTGCCCTCTGGCCCTTTCCCAGGAGCTCTCCTCCAGCACCGAGAGTATTGATAATTCATTCAGTTCCGTAAG .....................................................................................................((((....(((...(((....)))...))).))))............................................(((....)))............................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................TGAACCAAGTTGGAGCTAAATTCTTTGC.................................................................................................................. | 28 | 1 | 9.00 | 9.00 | 2.00 | 2.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATGAACCAAGTTGGAGCTAAATTCTT..................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGAACCAAGTTGGAGCTAAATTCTTTGCC................................................................................................................. | 29 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................AGCAGTTATGAACCAAGTTGGAGCTAAAT......................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATGAACCAAGTTGGAGCTAAATTCTTT.................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATGAACCAAGTTGGAGCTAAATTCT...................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...................................................................................................AAGCAGTTATGAACCAAGTTGGAGCTA............................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGAACCAAGTTGGAGCTAAATTCTTTG................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AAGCAGTTATGAACCAAGTTGGAGCTAAAT......................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................TGACTGTCTTTCACTAAAGCAGATATAGCTGC.............................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................TGTCTAGACTCTGGGCAACTGGCAGT..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................TGAACCAAGTTGGAGCTAAATTCTTT.................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGACTGTCTTTCgagc.............................................................................................................................................................................. | 16 | gagc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................TGAACCAAGTTGGAGCTAAATTCTTTGaa................................................................................................................. | 29 | aa | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| TGAGCTGGCCTTCCAACATCCCACAGATGTCTAGACTCTGGGCAACTGGCAGTTTCAGGCTGACTGTCTTTCACTAAAGCAGATATAGCTGCACAAGAGAAGCAGTTATGAACCAAGTTGGAGCTAAATTCTTTGCCTTTGTCTCTTTCCCTCCCCCCAACCCCACCCTTTCTTCCCCTTGCCCTCTGGCCCTTTCCCAGGAGCTCTCCTCCAGCACCGAGAGTATTGATAATTCATTCAGTTCCGTAAG .....................................................................................................((((....(((...(((....)))...))).))))............................................(((....)))............................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................GTTTCAGGCTGACTGTCTTTCACTAAAGCAGA....................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................................ATTGATAATTCATTCAGTTCCG.... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................TTTCAGGCTGACTGTCTTTCACTAAAGC.......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GTATTGATAATTCATTCAGTTCCGTA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |