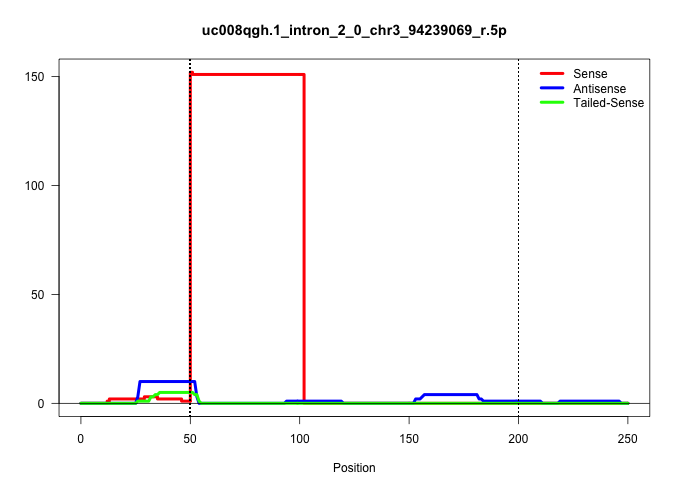
| Gene: Oaz3 | ID: uc008qgh.1_intron_2_0_chr3_94239069_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GGAGCACTGCACAGGAAAAAGACCACAGCCAGCTTAAAGAACTCTATTCAGTAAGTAAACTATGAACTGGTGATGTCACAGGGCAGCTCTGTGGTTTCTGAGAGAACATTAGCCAGTTGATAACCATAAAAATCAGAGTAGGTAGGACCCTGAGCTCATGTGTAAAATCTACTTGCTTTCTAGATGGATTCTGACCCAGAGATCTAAGTTATATAGCTGGTAGCCAACAGCAGGATAGGAAAAGTAGGTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTAAACTATGAACTGGTGATGTCACAGGGCAGCTCTGTGGTTTCTGAG.................................................................................................................................................... | 52 | 1 | 151.00 | 151.00 | 151.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CTTAAAGAACTCTATTCAGctg.................................................................................................................................................................................................... | 22 | ctg | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AGGAAAAAGACCACAGCCAGCTT....................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................AAGAACTCTATTCAGctg.................................................................................................................................................................................................... | 18 | ctg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TAAAGAACTCTATTCAGctgg................................................................................................................................................................................................... | 21 | ctgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............GGAAAAAGACCACAGCCAGCTTAAAGAACTCTA............................................................................................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................CAGCTTAAAGAACTCTATTCAG....................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................AGCCAGCTTAAAGAACTCTATTCAGc...................................................................................................................................................................................................... | 26 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAGCACTGCACAGGAAAAAGACCACAGCCAGCTTAAAGAACTCTATTCAGTAAGTAAACTATGAACTGGTGATGTCACAGGGCAGCTCTGTGGTTTCTGAGAGAACATTAGCCAGTTGATAACCATAAAAATCAGAGTAGGTAGGACCCTGAGCTCATGTGTAAAATCTACTTGCTTTCTAGATGGATTCTGACCCAGAGATCTAAGTTATATAGCTGGTAGCCAACAGCAGGATAGGAAAAGTAGGTA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................GCCAGCTTAAAGAACTCTATTCAGTA..................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................AGCCAGCTTAAAGAACTCTATTCAGTA..................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...........................GCCAGCTTAAAGAACTCTATTCAGTAA.................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................GCTCATGTGTAAAATCTACTTGCTTTCTA.................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTTCTGAGAGAACATTAGCCAGTTGA.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................AGCCAGCTTAAAGAACTCTATTCAGTAA.................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................ATGTGTAAAATCTACTTGCTTTCTAGA.................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GTAGCCAACAGCAGGATAGGAAAAGTAG... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GATGGATTCTGACCCAGAGATCTAAGTTA....................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CATGTGTAAAATCTACTTGCTTTCTA.................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |