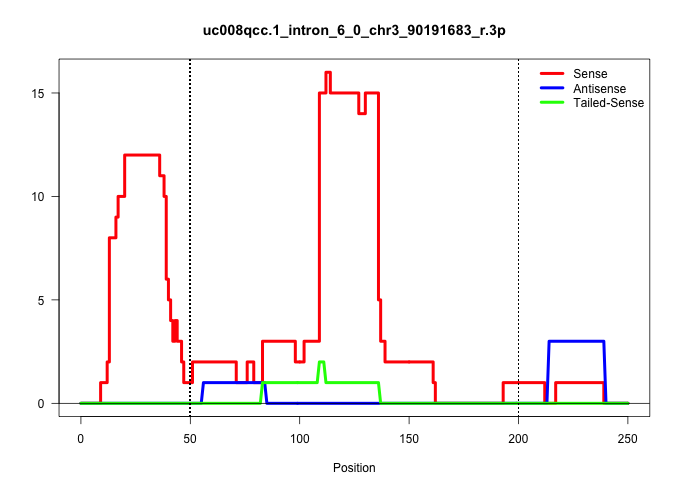
| Gene: Slc27a3 | ID: uc008qcc.1_intron_6_0_chr3_90191683_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TGTGGAGGCTGCATGGTTCAGAGGGAACAAAAGAGAAGGGTTGTATCAACCTAGAAAGGAATATTAGCCTCAGCAAACACGAGTAGGGAAGGACTTGGCCACTACAGCCTGCTAAAGACATGACTCCAGCAGGCGTCTGTAGTGAGCAATGGTGGCTGGAGTCTTCTCTTCACCCTCCCTTCTCTCCCGTTTCCGGGCAGGGGCCACCGTGGTGCTGAAACCCAAGTTCTCAGCTAGCCAGTTCTGGGAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TGCTAAAGACATGACTCCAGCAGGCGT.................................................................................................................. | 27 | 1 | 10.00 | 10.00 | 2.00 | 4.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............TGGTTCAGAGGGAACAAAAGAGAAGG................................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................TGCTAAAGACATGACTCCAGCAGGCGTC................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................CGGGCAGGGGCCACCGTGG...................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................TGTAGTGAGCAATGGTGGCTGGAGT........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................GAGGGAACAAAAGAGAAGGGTTGTAT............................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TAAAGACATGACTCCAGCAGGCGTCTG............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................ACACGAGTAGGGAAGGACTTGG........................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................AGGCGTCTGTAGTGAGCAATGGTGGCTGGAG......................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................TAAAGACATGACTCCAGCAGGCGTC................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TCAGAGGGAACAAAAGAGAAGGGTTGT.............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TATCAACCTAGAAAGGAATATTAGCCTC................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................TAGGGAAGGACTTGGCCACTACAGCCcgc.......................................................................................................................................... | 29 | cgc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGAAAGGAATATTAGCCTCAGCAAACA........................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGGTTCAGAGGGAACAAAAGAGAAG.................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................TTCAGAGGGAACAAAAGAGAAGGGTT................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGCTAAAGACATGACTCCAGCAGGCGga................................................................................................................. | 28 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGGAAGGACTTGGCCACTACAGCCTGC.......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AAACCCAAGTTCTCAGCTAGCC........... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............ATGGTTCAGAGGGAACAAAAGAGAAGGGT................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TGGTTCAGAGGGAACAAAAGAGAAGGG.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........TGCATGGTTCAGAGGGAACAAAAGAGA...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGGAAGGACTTGGCCACTACAGCCTGCTA........................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................GAGGGAACAAAAGAGAAGGGTTGTATC........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TACAGCCTGCTAAAGACATGACTCC........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTGGAGGCTGCATGGTTCAGAGGGAACAAAAGAGAAGGGTTGTATCAACCTAGAAAGGAATATTAGCCTCAGCAAACACGAGTAGGGAAGGACTTGGCCACTACAGCCTGCTAAAGACATGACTCCAGCAGGCGTCTGTAGTGAGCAATGGTGGCTGGAGTCTTCTCTTCACCCTCCCTTCTCTCCCGTTTCCGGGCAGGGGCCACCGTGGTGCTGAAACCCAAGTTCTCAGCTAGCCAGTTCTGGGAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................CTGAAACCCAAGTTCTCAGCTAGCCA.......... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGGAATATTAGCCTCAGCAAACACGAGTA..................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GGAAGGACTTGGCCACTACAGCCTGCTAa........................................................................................................................................ | 29 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |